
LINC00152 knockdown suppresses tumorigenesis in non-small cell lung cancer via sponging miR-16-5p
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and is known to be the leading cause of cancer-associated mortality worldwide (1). Surgery, radiotherapy and chemotherapy are currently the main treatment approaches for NSCLC. Although great efforts have been made over the past 20 years, the 5-year survival rate of patients with NSCLC remains low due to metastasis and recurrence (2). Therefore, novel treatment strategies for NSCLC are urgently required.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNA transcripts, ~200 nucleotides in length (3). The close association between lncRNAs and carcinogenesis has been previously reported (4,5). For example, lncRNA XIST-knockdown enhanced the chemosensitivity of NSCLC cells via inhibiting autophagy (6). Furthermore, LINC00152 is a lncRNA that was found to be upregulated in NSCLC (7); however, the underlying mechanism remains unclear and requires further investigation.
MicroRNAs (miRNAs) are a class of non-coding RNAs that regulate mRNA expression (8). It has been reported that miRNAs are involved in tumorigenesis (9). Kim et al. (10) indicated that miR-34a may act as an inhibitor of breast cancer via inhibition of Bcl-2. In addition, high expression of miR-21 was associated with a poor prognosis in patients with breast cancer (11). Therefore, miRNAs may serve as promising biomarkers in human malignancies. Additionally, another study suggested that miR-16-5p may be involved in the development of NSCLC (12), and LINC00152 could sponge miR-16-5p in glioma cells (13). However, the association between LINC00152 and miR-16-5p in NSCLC remains unclear.
The present study aimed to investigate the mechanism by which LINC00152 could regulate the growth of NSCLC cells, with the aim of shedding new light on the treatment of NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-59/rc).
Methods
Cell culture
The NSCLC cell lines (A549, NCI-H838, NCI-H3122 and NCI-H1975), and the human normal lung epithelial BEAS-2B cell line, were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences. Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; both Gibco; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) and 100 U/mL penicillin at 37 ℃.
Cell transfection
The small interfering RNA (siRNA) targeting LINC00152 (si-LINC00152; 10 nM) and the corresponding negative control siRNA (si-NC) were purchased from Guangzhou RiboBio Co., Ltd. and transfected into NSCLC cells (5×103) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA). The transfection efficiency was evaluated using reverse transcription‑quantitative polymerase chain reaction (RT-qPCR). The sequences of the siRNAs were as follows: For si-NC, 5'-UUCUCCGAACGUGUCACGUTT-3'; and for si-LINC00152, 5'-GGGTTACGATTGCCCAGAT-3'.
For miR-16-5p transfection, A549 cells were transfected with miR-16-5p inhibitor or inhibitor-control (NC) using Lipofectamine® 2000(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA), as previously described (14). miR-16-5p inhibitor and the corresponding NC were purchased from Shanghai GenePharma Co., Ltd., Shanghai, China. After 24 h of transfection, transfected cells were used for subsequent experimentation.
BCL2 like 2 (BCL2L2)-overexpression
NSCLC cells were transfected with the pcDNA3.1 vector (NC) or pcDNA3.1-BCL2L2 (BCL2L2-overexpression) at 37 ℃ for 24 h using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA). The pcDNA3.1 vector and pcDNA-BCL2L2 were purchased from Shanghai GenePharma Co., Ltd., Shanghai, China. The cell supernatant was collected 48 h following transfection.
Bioinformatics analysis
The association between LINC00152 and the prognosis of lung adenocarcinoma (LUAD) was analyzed using The Cancer Genome Atlas (TCGA) and Gene expression profiling interactive analysis (GEPIA) databases (http://gepia.cancer-pku.cn/index.html) (15).
RT-qPCR analysis
Total RNA was extracted from NSCLC cells using TRIzol® reagent and reverse-transcribed into cDNA using the PrimeScript RT-PCR kit (both Takara Biotechnology Co., Ltd. Shiga, Japan). The temperature and duration of reverse-transcription were as follows: 37 ℃ for 60 min and 85 ℃ for 5 min. The mRNA expression levels of each gene were determined in triplicate using the SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd., Shiga, Japan) kit on a Real-Time PCR System (Applied Biosystems 7500; Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) and normalized to β-actin. The amplification protocol was set as follows: Incubation at 95 ℃ for 5 min, followed by 40 cycles at 95 ℃ for 10 sec and 60 ℃ for 30 sec. The primers for LINC00152, miR-16-5p, β-actin and U6 were obtained from Shanghai GenePharma Co., Ltd., Shanghai, China, and the primer sequences were as follows: LINC00152 forward, 5'-TTTTTTTCCTTCTTAGTCGTGTGTA-3' and reverse, 5'-AAATTGACATTCCAGAC-3'; miR-16-5p forward, 5'-TAGCAGCACGTAAATATTGGCG-3' and reverse, 5'-TGCGTGTCGTGGAGTC-3'; β-actin forward, 5'-AGCGAGCATCCCCCAAAGTT-3' and reverse 5'-GGGCACGAAGGCTCATCATT-3'; and U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'. The 2−ΔΔCq method was used for quantification (16).
CCK-8 assay
The NSCLC cell viability in each group was evaluated using a CCK-8 assay kit (Beyotime Institute of Biotechnology). The optical density values at a wavelength of 450 nm were measured using a microplate reader to determine cell viability.
Cell apoptosis analysis
For the analysis of cell apoptosis, the Annexin V Apoptosis Detection kit (BD Biosciences) was used according to the manufacturer’s protocols. The cells were analyzed by flow cytometry (BD Biosciences). The results were analyzed using a flow cytometer (FACS; BD Biosciences) with FlowJo software (v10.6.2; BD Biosciences).
Dual-luciferase reporter assay
The miRNAs targeting LINC00152 were predicted using the starBase online software (version 2.0; http://starbase.sysu.edu.cn/index.php). miR-16-5p was predicted as the miRNA directly targeting LINC00152. Subsequently, the protein target of miR-16-5p was predicted using two online databases, namely TargetScan (http://www.targetscan.org/vert_72/) and miRDB (http://www.mirdb.org/). The analysis predicted that BCL2L2 could be the putative protein target of miR-16-5p. A dual-luciferase reporter assay was performed to validate the aforementioned findings. The wild-type (WT) and mutant (MUT) LINC00152 were sub-cloned into the pmiR-RB-Report™ dual-luciferase reporter vectors (Guangzhou RiboBio Co., Ltd., Guangzhou, China). Furthermore, NSCLC cell lines were co-transfected with miR-16-5p mimics (Guangzhou RiboBio Co., Ltd., Guangzhou, China) or control vector and the WT- or MUT-LINC00152 reporter plasmid using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) at 37 ℃ for 48 h according to the manufacturer’s protocols. The luciferase activity was measured by a Dual-luciferase Assay System (Promega Corporation, Madison, Wisconsin, USA). The interaction between BCL2L2 and miR-16-5p was also verified using a dual-luciferase reporter assay. The positions in the sequence were presented in http://www.mirdb.org/cgi-bin/target_detail.cgi?targetID=1788842. After 48 h of transfection, the relative luciferase activity was detected using the Dual-Luciferase Reporter kit (Promega Corporation, Madison, Wisconsin, USA). The data were normalized to Renilla luciferase activity.
Western blot analysis
Cell lysates were prepared using a RIPA lysis buffer (Shanghai GenePharma Co., Ltd., Shanghai, China) and proteins were quantified with a BCA protein assay kit (Beyotime Institute of Biotechnology). Equal amounts (20 µg) of protein extracts from each group were separated by 10% SDS-PAGE and were then transferred onto a PVDF membrane. Following blocking with 10% FBS for 1 h at room temperature, the membranes were first incubated with primary antibodies against BCL2L2 (dilution, 1:1,000; cat. no. ab190952), Bax (dilution, 1:1,000; cat. no. ab32503), Bcl-2 (dilution, 1:1,000; cat. no. ab182858), and caspase 3 (dilution, 1:1,000; cat. no. ab49822), and then with horseradish peroxidase-labeled goat anti-rabbit secondary antibodies (dilution, 1:5,000; cat. no. ab7090; all from Abcam). GAPDH was utilized as an endogenous control. Enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) was used to visualize the protein bands.
Transwell assay
A 24-well Transwell chamber with an 8-µm pore membrane (Corning Incorporated) was utilized for the Transwell invasion assay. For the invasion assay, the upper chamber was pre-treated with 50 µL Matrigel (12.5 mg/L). NSCLC cells (4×104) were suspended in 200 µL DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) without FBS. Cell culture medium supplemented with 20% FBS was added into the lower chamber to serve as a chemoattractant. Following incubation for 24 h, the invaded cells were fixed with 4% paraformaldehyde for 10 min at room temperature and stained with 0.25% crystal violet for 5 min at room temperature. Images were captured under a light microscope (magnification, ×200; Olympus Corporation, Tokyo, Japan).
Statistical analysis
All data are expressed as the mean± standard deviation. Differences between groups were compared using one-way analysis of variance, followed by Tukey’s post hoc test, with GraphPad Prism 8 (GraphPad Software Inc., California, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
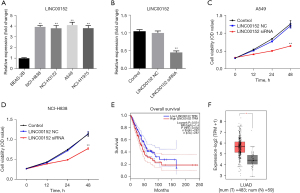
LINC00152-silencing significantly attenuates the proliferation of NSCLC cells. To investigate the biological effect of LINC00152 on NSCLC, RT-qPCR analysis was performed. As shown in Figure 1A, LINC00152 was notably upregulated in NSCLC cells. Since A549 and NCI-H838 were more sensitive to LINC00152 expression, these two cell lines were selected for further analysis. In addition, NSCLC cell lines were efficiently transfected with si-LINC00152 (Figure 1B). Subsequently, a CCK-8 assay was performed. The results revealed that LINC00152-silencing notably attenuated NSCLC cell proliferation (Figure 1C,1D). Since the A549 cells were more susceptible to si-LINC00152, A549 cells were selected for subsequent experiments. Additionally, TCGA data analysis revealed that LINC00152 was associated with a poor prognosis in NSCLC (Figure 1E), and the expression of LINC00152 in LUAD tissues was higher than that in adjacent normal tissues (Figure 1F). These findings indicated that LINC00152 was upregulated in NSCLC.
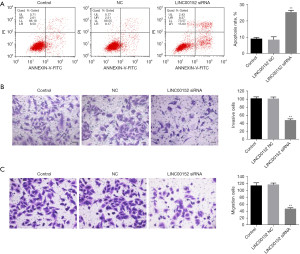
Knockdown of LINC00152 notably increases apoptosis and inhibits the invasion of NSCLC cells. Subsequently, flow cytometry was performed to measure cell apoptosis. As expected, downregulation of LINC00152 increased the apoptosis of A549 and NCI-H838 cells (Figure 2A, Figures S1A,S1B). Similarly, NSCLC cell migration and invasion were notably decreased following transfection of A549 cells with si-LINC00152 siRNA (Figure 2B,2C). Taken together, these findings demonstrated that LINC00152-knockdown could notably increase apoptosis and decrease the migration and invasion ability of NSCLC cells.
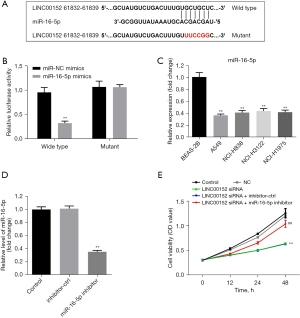
LINC00152 inhibits the tumorigenesis of NSCLC via miR-16-5p. To investigate the mechanism underlying the effect of LINC00152 on NSCLC tumorigenesis, the Starbase database was used. As shown in Figure 3A, miR-16-5p could be the downstream miRNA target of LINC00152. Furthermore, the relative luciferase activity of WT-LINC00152 was significantly decreased in the presence of miR-16-5p mimics (Figure 3B). In addition, the RT-qPCR results demonstrated that miR-16-5p was markedly downregulated in NSCLC cells (Figure 3C). Furthermore, the miR-16-5p inhibitor was stably transfected into NSCLC cells (Figure 3D), and miR-16-5p downregulation partially rescued the antiproliferative effect of si-LINC00152 on A549 cells (Figure 3E). The aforementioned findings suggested that LINC00152 could attenuate the progression of NSCLC via downregulation of miR-16-5p.
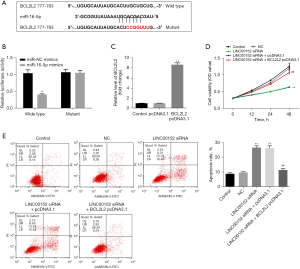
LINC00152 indirectly targets BCL2L2. To identify the targets of miR-16-5p, the TargetScan and miRDB databases were used. The results indicated that miR-16-5p could directly target BCL2L2 (Figure 4A). In addition, the dual-luciferase reporter assay further confirmed these finding (Figure 4B). Meanwhile, the expression of BCL2L2 in NSCLC cells was significantly increased in the presence of pcDNA3.1-BCL2L2 (Figure 4C). Furthermore, BCL2L2-overexpression significantly reversed the inhibitory effect of si-LINC00152 on NSCLC cell proliferation (Figure 4D). Consistently, the apoptotic effect of si-LINC00152 on NSCLC cells was markedly inhibited following BCL2L2-overexpression (Figure 4E). In summary, LINC00152 could indirectly target BCL2L2.
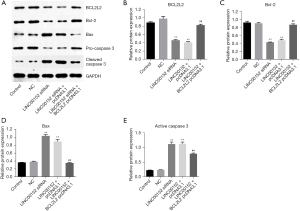
Overexpression of BCL2L2 significantly attenuates the effect of LINC00152-silencing on the expression of apoptotic proteins in NSCLC cells. Finally, to further investigate the effect of LINC00152 on NSCLC in vitro, Western blot analysis was performed. As expected, the expression levels of BCL2L2 and Bcl-2 in NSCLC cells were significantly decreased by LINC00152-knockdown. This effect was notably reversed by BCL2L2-overexpression (Figure 5A-5C). By contrast, LINC00152-knockdown notably enhanced the protein expression levels of Bax and caspase-3. However, the effect of si-LINC00152 on the two proteins was markedly suppressed following BCL2L2-overexpression (Figure 5A,5D,5E). The aforementioned results suggested that BCL2L2 overexpression could significantly attenuate the effect of LINC00152-silencing on the expression of apoptotic proteins in NSCLC cells.
Discussion
It has been previously reported that LINC00152 is involved in tumorigenesis (17-19). For example, a study demonstrated that LINC00152 may serve a key role in the progression of colorectal cancer (20). These findings suggested that LINC00152 could act as an oncogene in several types of cancer. In addition, it has been reported that LINC00152 is upregulated in NSCLC, and that LINC00152 could promote lung adenocarcinoma proliferation via interacting with EZH2 and inhibiting IL24 transcription (7,21). The findings of the present study were consistent with these previous results, further verifying that LINC00152 may serve an important role in the tumorigenesis of NSCLC.
miRNAs have multiple biological functions (22). The present study revealed that LINC00152 was associated with miR-16-5p. Several previous studies have reported that miR-16-5p is involved in different types of malignant tumors and that it may act as a tumor suppressor (22-24). A previous study demonstrated that miR-16-5p could inhibit NSCLC development (12). In addition, Chen et al. (13) reported that LINC00152 could directly bind with miR-16-5p in glioma. The results of the present study were consistent with those of previous studies. However, another study suggested that LINC00152 may regulate the progression of esophageal squamous cell carcinoma via regulating miR-107 (19). This difference may be due to the different tumor type.
miRNAs exert their biological functions mainly via regulating the expression of their target mRNAs (9). In the present study, BCL2L2 was revealed to be directly targeted by miR-16-5p. BCL2L2 has been considered to act as an inhibitory factor during the apoptosis of cells, similarly to Bcl-2 (25). Liu et al. (26) confirmed that miR-93 may alleviate the proliferation and induce the apoptosis of trophoblast cells via targeting BCL2L2 in recurrent spontaneous abortion. miR-126-5p has been considered to regulate the tumorigenesis of ovarian cancer via targeting BCL2L2 (27). According to the results of a previous study (13), LINC00152 may promote the tumorigenesis of glioma through sponging miR-16. In addition, BCLCL2 was found to be the direct target of miR-16 in the present study. Therefore, it can be suggested that LINC00152 regulates the expression of BCLCL2 through sponging BCL2L2. However, Yin et al. (28) indicated that PIK3R1 and RAF1 were directly targeted by miR-16-5p. These differences may be attributed to the different types of diseases investigated.
Additionally, the present study demonstrated that LINCC00152-silencing may promote the apoptosis of NSCLC cells. Furthermore, LINC00152-silencing significantly increased the protein expression levels of Bax and caspase-3 and downregulated those of Bcl-2. Bax, caspase-3 and Bcl-2 are considered to be key mediators of cell apoptosis in several diseases (2,29,30). Emerging evidence has supported that Bax and caspase-3 are pro-apoptotic proteins (31), while Bcl-2 upregulation inhibits cell apoptosis (32). Furthermore, the present study demonstrated that LINC00152-knockdown induced NSCLC cell apoptosis via upregulating Bax and caspase-3, and downregulating Bcl-2.
However, the present study is limited by the fact that the effect of LINC00152 siRNA on NSCLC cell proliferation remains to be further verified. Therefore, further investigations are required in the future.
In conclusion, the results of the present study demonstrated that LINC00152-knockdown inhibited the development of NSCLC via regulating the miR-16-5p/BCL2L2 axis, thereby providing novel insight into the treatment of NSCLC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-59/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-59/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-59/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Song P, Zhang X, Yang D, et al. Single-center study to determine the safety and efficacy of CT-707 in Chinese patients with advanced anaplastic lymphoma kinase-rearranged non-small-cell lung cancer. Thorac Cancer 2020;11:1216-23. [Crossref] [PubMed]
- Chi F, Wang Z, Li Y, et al. Knockdown of GINS2 inhibits proliferation and promotes apoptosis through the p53/GADD45A pathway in non-small-cell lung cancer. Biosci Rep 2020;40:BSR20193949. [Crossref] [PubMed]
- Zhou M, Bian Z, Liu B, et al. Long noncoding RNA MCM3AP-AS1 enhances cell proliferation and metastasis in colorectal cancer by regulating miR-193a-5p/SENP1. Cancer Med 2021;10:2470-81. [Crossref] [PubMed]
- Chen G, Jin X, Luo D, et al. β-Adrenoceptor regulates contraction and inflammatory cytokine expression of human bladder smooth muscle cells via autophagy under pathological hydrostatic pressure. Neurourol Urodyn 2020;39:2128-38. [Crossref] [PubMed]
- Geng C, Li Y, Guo F, et al. RNA sequencing analysis of long non-coding RNA expression in ocular posterior poles of guinea pig myopia models. Mol Vis 2020;26:117-34. [PubMed]
- Tian LJ, Wu YP, Wang D, et al. Upregulation of Long Noncoding RNA (lncRNA) X-Inactive Specific Transcript (XIST) is Associated with Cisplatin Resistance in Non-Small Cell Lung Cancer (NSCLC) by Downregulating MicroRNA-144-3p. Med Sci Monit 2019;25:8095-104. [Crossref] [PubMed]
- Yu H, Li SB. Role of LINC00152 in non-small cell lung cancer. J Zhejiang Univ Sci B 2020;21:179-91. [Crossref] [PubMed]
- Tseng KC, Chiang-Hsieh YF, Pai H, et al. sRIS: A Small RNA Illustration System for Plant Next-Generation Sequencing Data Analysis. Plant Cell Physiol 2020;61:1204-12. [Crossref] [PubMed]
- Xie W, Luo J, Pan C, et al. SG-LSTM-FRAME: a computational frame using sequence and geometrical information via LSTM to predict miRNA-gene associations. Brief Bioinform 2021;22:2032-42. [Crossref] [PubMed]
- Kim D, Lee J, Kang J, et al. Notch1 in Tumor Microvascular Endothelial Cells and Tumoral miR-34a as Prognostic Markers in Locally Advanced Triple-Negative Breast Cancer. J Breast Cancer 2019;22:562-78. [Crossref] [PubMed]
- Kundaktepe BP, Sozer V, Papila C, et al. Associations Between miRNAs and Two Different Cancers: Breast and Colon. Cancer Manag Res 2020;12:871-9. [Crossref] [PubMed]
- Wang Q, Chen Y, Lu H, et al. Quercetin radiosensitizes non-small cell lung cancer cells through the regulation of miR-16-5p/WEE1 axis. IUBMB Life 2020;72:1012-22. [Crossref] [PubMed]
- Chen X, Li D, Gao Y, et al. Long Intergenic Noncoding RNA 00152 Promotes Glioma Cell Proliferation and Invasion by Interacting with MiR-16. Cell Physiol Biochem 2018;46:1055-64. [Crossref] [PubMed]
- Zhou X, Zhang Z, Liang X. Regulatory Network Analysis to Reveal Important miRNAs and Genes in Non-Small Cell Lung Cancer. Cell J 2020;21:459-66. [PubMed]
- Sun CC, Li SJ, Hu W, et al. Comprehensive Analysis of the Expression and Prognosis for E2Fs in Human Breast Cancer. Mol Ther 2019;27:1153-65. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Li SQ, Chen Q, Qin HX, et al. Long Intergenic Nonprotein Coding RNA 0152 Promotes Hepatocellular Carcinoma Progression by Regulating Phosphatidylinositol 3-Kinase/Akt/Mammalian Target of Rapamycin Signaling Pathway through miR-139/PIK3CA. Am J Pathol 2020;190:1095-107. [Crossref] [PubMed]
- Seo D, Kim D, Kim W. Long non-coding RNA linc00152 acting as a promising oncogene in cancer progression. Genomics Inform 2019;17:e36. [Crossref] [PubMed]
- Zhou Z, Huang F. Long Non-Coding RNA LINC00152 Regulates Cell Proliferation, Migration And Invasion In Esophageal Squamous Cell Carcinoma Via miR-107/Rab10 Axis. Onco Targets Ther 2019;12:8553-67. [Crossref] [PubMed]
- Nalin N, Al Dhanhani A, AlBawardi A, et al. Effect of angiotensin II on diabetic glomerular hyperpermeability: an in vivo permeability study in rats. Am J Physiol Renal Physiol 2020;319:F833-8. [Crossref] [PubMed]
- Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer 2017;16:17. [Crossref] [PubMed]
- Casabonne D, Benavente Y, Seifert J, et al. Serum levels of hsa-miR-16-5p, hsa-miR-29a-3p, hsa-miR-150-5p, hsa-miR-155-5p and hsa-miR-223-3p and subsequent risk of chronic lymphocytic leukemia in the EPIC study. Int J Cancer 2020;147:1315-24. [Crossref] [PubMed]
- Xie F, Xie G, Sun Q. Long Noncoding RNA DLX6-AS1 Promotes the Progression in Cervical Cancer by Targeting miR-16-5p/ARPP19 Axis. Cancer Biother Radiopharm 2020;35:129-36. [Crossref] [PubMed]
- Duran-Sanchon S, Vila-Navarro E, Marcuello M, et al. Validation of miR-1228-3p as Housekeeping for MicroRNA Analysis in Liquid Biopsies from Colorectal Cancer Patients. Biomolecules 2019;10:16. [Crossref] [PubMed]
- Kacı FN, Kiraz Y, Çekdemir D, et al. Synergistic Apoptotic Effects of Bortezomib and Methylstat on Multiple Myeloma Cells. Arch Med Res 2020;51:187-93. [Crossref] [PubMed]
- Liu HN, Tang XM, Wang XQ, et al. MiR-93 Inhibits Trophoblast Cell Proliferation and Promotes Cell Apoptosis by Targeting BCL2L2 in Recurrent Spontaneous Abortion. Reprod Sci 2020;27:152-62. [Crossref] [PubMed]
- Stover EH, Baco MB, Cohen O, et al. Pooled Genomic Screens Identify Anti-apoptotic Genes as Targetable Mediators of Chemotherapy Resistance in Ovarian Cancer. Mol Cancer Res 2019;17:2281-93. [Crossref] [PubMed]
- Yin K, Cui Y, Qu Y, et al. Hydrogen sulfide upregulates miR-16-5p targeting PiK3R1 and RAF1 to inhibit neutrophil extracellular trap formation in chickens. Ecotoxicol Environ Saf 2020;194:110412. [Crossref] [PubMed]
- Bao J, Chen Z, Xu L, et al. Rapamycin protects chondrocytes against IL-18-induced apoptosis and ameliorates rat osteoarthritis. Aging (Albany NY) 2020;12:5152-67. [Crossref] [PubMed]
- Qin H, Zhang LL, Xiong XL, et al. Li-Dan-He-Ji Improves Infantile Cholestasis Hepatopathy Through Inhibiting Calcium-Sensing Receptor-Mediated Hepatocyte Apoptosis. Front Pharmacol 2020;11:156. [Crossref] [PubMed]
- Brokatzky D, Kretz O, Häcker G. Apoptosis Functions in Defense against Infection of Mammalian Cells with Environmental Chlamydiae. Infect Immun 2020;88:e00851-19. [Crossref] [PubMed]
- Yirong C, Shengchen W, Jiaxin S, et al. DEHP induces neutrophil extracellular traps formation and apoptosis in carp isolated from carp blood via promotion of ROS burst and autophagy. Environ Pollut 2020;262:114295. [Crossref] [PubMed]


