Integrated gene expression profiling analysis reveals SERPINA3, FCN3, FREM1, MNS1 as candidate biomarkers in heart failure and their correlation with immune infiltration
Introduction
As a complex clinical syndrome, heart failure (HF) has become the most common cause of adult hospitalization in developed countries (1) HF is generally regarded as an irreversible manifestation of long-term heart disease. HF is most often caused by ischemic heart disease (IHD) or dilated cardiomyopathy (DCM) (HF). Cardiac insufficiency is present in different syndromes, however, they do not have the precise symptoms and indications of a specific condition. The most common cause of persistent congestive HF is DCM, which is characterized by nonischemic left ventricular enlargement and alterations in cardiac structure and function. HF develops as a result of dilatation of the heart’s chambers, which is brought on by IHD’s chronic ventricular remodeling. Currently, therapeutic methods are limited to delaying the development of the disease and treating symptoms of volume congestion. Chronic HF may lead to frequent hospitalization and long-term treatment with cardiac assistance devices or even a heart transplant, based on the clinical course of the disease. Myocardial gene expression disorder has been shown to be the mediator of common end-stage diseases. In the end-stage, the heart responds to multiple developmental pathways. Although the central regulatory mechanism leading to cardiac transcriptional reprogramming remains unclear, epigenetic contributions may exist. Therefore, regardless of the etiology, we are committed to identifying the common molecular mechanism of the pathogenesis of end-stage cerebral infarction.
HF is characterized by overall changes in gene expression in the myocardium, including the reactivation of developmental pathways (2). Family sequencing analysis and genome-wide association study (GWAS) (3) have identified numerous single-gene mutations and gene variations, respectively, which are sufficient to make HF diagnosis easily . Although environmental stimulation alters cardiac gene expression, the molecular mechanism remains unknown. Ecological variables such as diet and exercise have been thought to be regulators of genetic risk. Epigenetics is well suited to explain how changes in the cardiac milieu cause persistent but changing transcriptional responses (4). Any number of ischemia and nonischemic pathophysiological events may have an impact on the myocardium. In the majority of cases, irreversible HF follows acute ischemia damage or a gradual decline in cardiac function as a result of numerous clinicopathological factors. Ischemic injury is the main cause of myocardial injury. Tissue-resident immune and non-immune cells are activated when damaged and necrotic cardiomyocytes die as a result of an ischemia shock to the myocardium. The expansion of neutrophil and macrophage populations results in the release of cytokines and growth factors that promote the creation of highly vascularized granulation tissue (i.e., connective tissue and new vasculature). Adhesion molecule expression and the corresponding immune cell infiltration might improve disease diagnostics and improve treatment targets via a better knowledge of their expression (5).
In this study, we initially obtained HF biochip data from the Gene Expression Omnibus (GEO) database (6) to analyze differentially-expressed genes (DEGs). Next, a machine learning algorithm was used to further screen and identify the diagnostic factors of HF (7). We present the following article in accordance with the STARD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-22/rc).
Methods
Data download
GSE 76701 and Series Matrix File data files from the NCBI (The National Center for Biotechnology Information advances science and health by providing access to biomedical and genomic information.) GEO public database (http://www.ncbi.nlm.nih.gov/geo/) was downloaded using the R package “GEOquery” (version 3.6.1, http://r-project.org/) (8) and the “GEOquery” package of R software (version 3.6.1, http://r-project.org/) (9). The study was conducted in accordance with the Helsinki Declaration (as revised in 2013).
Data preprocessing
A comparison between the Series Matrix and GSE76701 of the NCBI GEO public database was performed using the annotation platform GPL570 (Genome). A total of eight transcriptome data sets were obtained, including four from the normal group and four from the illness group. GSE57338 and Series Matrix File data files were downloaded with the annotation platform GPL11532, derived from myocardial array data, with a total of 231 transcriptomic data sets, including both normal (n=136) and disease (n=95) groups, which were used for data correction between microarrays with the System Verilog assertion (SVA) algorithm, in order to explore the differences between disease-related molecular mechanisms. We also downloaded the GSE16499 dataset (annotation platform: GPL5175), with a total 30 sets of transcriptome data, including both normal (n=15) and disease (n=15) groups for subsequent validation.
Functional correlation analysis
We used the Metascape databases (https://www.metascape.org) to obtain the biological functions and signaling pathways involved in disease development. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on specific genes. A minimum overlap ≥3 and P≤0.01 were considered statistically significant.
Screening and verification of diagnostic factors
We used least absolute shrinkage and selection operator (LASSO) logistic regression and Boruta algorithms for feature selection of diagnostic factors of disease. The LASSO algorithm was applied using the “glmnet” R package. Furthermore, Boruta is a feature selection algorithm that randomly disrupts the order of each real feature and evaluates the importance of each feature, iteratively removing features with low correlation to find the best variables. A total of 500 trees were built to further identify the diagnostic value of the biofactors for the illness using the “Boruta” program on a selection of features.
Evaluation of immune cell infiltration
We used ssGSEA to quantify the level of immune cell infiltration in each sample and evaluate the effect of genes on immune infiltration. Sixty-four immunological and stromal cell types were studied using xCell (http://xCell.ucsf.edu/) and thorough in silico studies that were also compared with immunophenotyping. The link between gene expression and the content of the immune cells was examined.
Gene set differentiation analysis (GSVA)
Gene enrichment of transcriptome gene sets may be assessed using the non-parametric unmonitored GSVA approach. GSVA converts the gene level change into a pathway level change and then evaluates the sample’s biological function by synthetically scoring the relevant gene set. The GSVA method was used to thoroughly assess each gene set to assess the possible physical function alterations in various samples downloaded from the Molecular signatures database (v7.0 edition).
Statistical analysis
The statistical analysis is carried out in R language (version 3.6). All statistical tests were bilateral. P<0.05 was statistically significant.
PPI network analysis and hub gene searching
For DEGs, we used the Search Tool for the Retrieval of Interacting Genes (STRING) to anticipate functional protein interactions. Through Molecular Complex Detection (MCODE) analysis, we can identify the most strongly linked cluster from the PPI network. The genes in the retrieved network with a high Maximal Clique Centrality (MCC) score determined by CytoHubba were designated as hub genes.
ROC curve analysis
It is possible to compress and preserve critical variables using the glmnet (Binomial LASSO) tool available in the R programming language. To calculate the area under the curve (AUC) and construct the receiver operating characteristic (ROC) curves for each cohort independently, we utilized PRISM8.0 (Network Analysis Tools) to perform a regression of the AUC. As a result, we looked at the potential of hub genes to forecast via the use of the AUC value.
Results
Data preprocessing and DEGs screening
We downloaded the GSE76701 and GSE57338 HF-related data sets from the GEO database, with a total of 239 groups of patients, including both normal (n=140) and disease (n=99) groups. The SVA algorithm was used to correct the chip, and a principal component analysis (PCA) chart was used to show the difference before and after correction. The results showed that the batch effect between chips was eliminated after SVA correction (Figure 1). We further used the limma package to calculate the differential genes between the two groups. The differential gene screening conditions were P<0.05 and |log2fc| >1. A total of 43 differential genes were screened out, including 24 up-regulated genes and 19 down-regulated genes (Figure 1C).

Functional correlation analysis
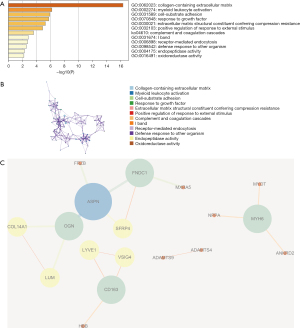
We then analyzed the pathways of differential genes. We observed that the primary enrichment for differential genes is the collagen-containing extracellular matrix, myeloid leukocyte activation, cell-substrate adhesion, growth factor response, and extracellular matrix structural constituent conferring compression resistance (Figure 2). An interaction diagram between the genes is shown in Figure 2C.
Screening and verification of diagnostic factors
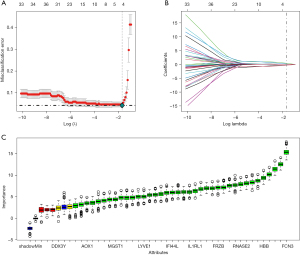
LASSO regression and Boruta feature selection algorithms were employed to identify essential genes in the differential gene, and distinctive HF genes were screened out using these methods. Four critical genes associated with HF were discovered using the LASSO logistic regression technique (Figure 3A,3B). The Boruta algorithm identified 38 significant genes related to HF (Figure 3C).
The core genes were verified by external data sets
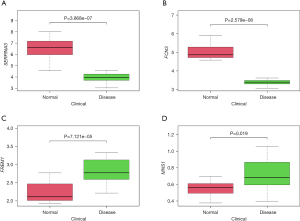
Four key genes were selected after the intersection: SERPINA3, FCN3, FREM1, and MNS1. We further downloaded the GSE16499 HF data sets from the GEO public database and corresponding clinical data. Through validation of key gene differential analysis, where FCN3 (P=2.579e−08), FREM1 (P=7.121e−05), MNS1 (P=0.019), and SERPINA3 (P=3.868,07), we determined that all had differential significance (Figure 4).
ROC diagnostic efficacy of core genes
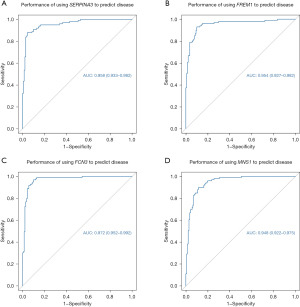
The ROC curve was validated by diagnostic efficacy; the higher the AUC, the better diagnostic efficacy. The results showed that the AUC values of the four core genes were FCN3 [0.972 (0.952–0.992)], FREM1 [0.954 (0.927–0.982)], MNS1 [0.948 (0.922–0.975)], and SERPINA3 [0.958 (0.933–0.982)] (Figure 5).
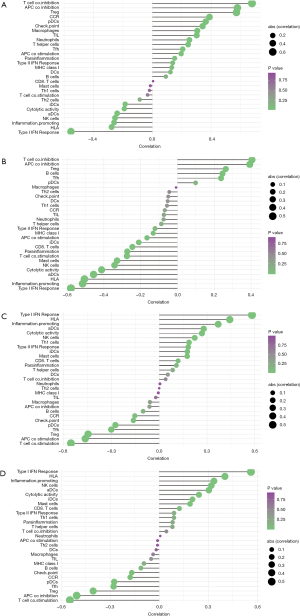
Correlation analysis between SERPINA3, FCN3, FREM1, MNS1 and infiltrating immune cells
Disease detection and therapy are made much more sensitive when the milieu is in place. The microenvironment includes immune cells, extracellular matrix, and growth factors, as well as other physical and chemical properties. By analyzing the relationship between core genes and immune infiltration in the HF data set, the potential molecular mechanism of core genes affecting the progression of HF was further explored. All four genes exhibited a strong correlation with immune cell content (Figure 6), and the results were in line with expectations. With a P value cutoff of 0.05, the estimated proportions of immune and stromal cell types were derived for each cardiac tissue sample using xCell and the Wilcoxon technique for variance assessment. Cells were categorized as lymphoid [Tregs, B cells, Th1 cells, Th2 cells, NK cells, NK T cells, activated dendritic cells (aDCs), CD4+ effector memory T cells (Tem), CD8+ naive T cells, CD8+ T cells, macrophage M1, macrophage M2, immature dendritic cells (iDCs), and others (epithelial cells, sebocytes, keratinocytes, mesangial cells, hepatocytes, melanocytes, astrocytes, neurons)] (10).
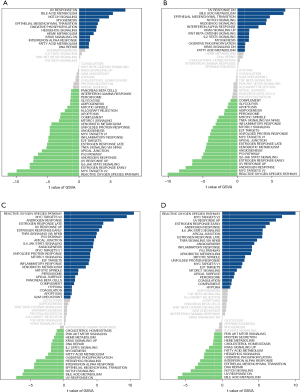
GSVA analysis of key genes
To explore the potential molecular mechanism of core genes affecting the progression of HF, we studied the specific signaling pathways involved in the four core genes. The GSVA results showed that the high expression of FREM1 and MNS1 was involved in bile acid, fatty acid, and heme metabolism (11), suggesting that the core gene affects the progression of HF by regulating metabolism. Meanwhile, the high expression of FCN3 and SERPINA3 was found to be related to xenobiotic metabolism, inflammatory response, and adipogenesis (Figure 7).
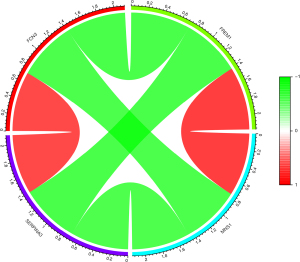
Expression control of core genes correlation diagram
We used the “circlize” and “corrplot” packages to draw interaction maps expressing positively and negatively correlated core genes, where green represents a negative correlation and red represents a positive correlation (Figure 8).
Discussion
Congestive HF (CHF) (12) is the ultimate common outcome of multiple hearts diseases. Increases in the incidence of cardiovascular disorders such as hypertension, coronary heart disease, and stroke are associated with the aging of the population. The incidence of the disease is growing, and in the occurrence of symptoms, the 5-year survival rate is fewer than half of those who are diagnosed. The mortality rate is comparable to some common malignancies. Therefore, the treatment and drug development for HF are crucial. The pathogenesis, initiation, and progression of CHF are complex (13). Cardiac-respiratory, cardiopulmonary, cardiac-circulation, and cardioneurohumoral patterns have been seen in the HF hypothesis. As these theories evolve, CHF drug development and treatment strategies are also changing, especially neuroendocrine blockers. The curative effect of HF has been considerably improved. Recent research (14) has shown that endocrine blockers have reached their limits (ceiling effect). As of yet, no new approaches or medications have been discovered for treating HF or reducing mortality. Therefore, identifying new therapeutic drugs and methods is urgently needed. Therapeutic target intervention of HF has become an essential approach to the treatment of HF (15). In recent years, with the deepening understanding of the pathophysiological process of HF, new potential drug intervention targets have emerged as a cure for HF. At the same time new drug research and development to provide ideas (16).
We used GSEA to analyze the KEGG pathways associated with immunological infiltration. There is strong evidence that the core gene, FREM1 and MNS1, impacts the course of HF by controlling the metabolism of bile, fat, and heme, according to GSVA data. Adipogenesis, xenobiotic metabolism, and inflammation were all associated with the elevated expression of FCN3 and SERPINA3. FCN3 is linked to the development of HF. MNS1, SLCO4A1, and FREM1 were added in a support vector machine-based risk prediction model. In our training and validation cohorts, these conventional biomarkers performed well in predicting the risk of HF.
With the accumulation of large-scale omics data, experimental methods alone can no longer satisfy data analysis and drug target discovery (17). It is necessary to develop effective bioinformatics methods to store, analyze, process, and integrate multi-omics data, in order to improve drug target discovery and verification efficiency. At present, bioinformatics methods have been successfully used in all aspects of drug target discovery, which has provided essential contributions to storing disease-related medical data, discovering a large number of potential drug targets, revealing the mechanism of drug action, and evaluating the drug availability of action targets (18). Furthermore, it is helpful to design more targeted biological experiments and promote the development of modern drugs. The benefits of bioinformatics (19) compared with other methods in predicting potential drug targets are as follows: (I) it is not limited to specific technologies or types of information and is particularly suited to integrating differing information into a comprehensive system for evaluating the performance of potential drug targets; (II) networked drug discovery target platforms improve overall drug goal screening and common goal identification; and (III) the process of drug targeting and the effect on the entire system on a computer may be correctly simulated with the accumulation of dynamically precise biological Spatio-temporal data to significantly improve drug efficiency development (20).
Traditional HF biofactors, such as Brain Natriuretic Peptide (BNP) or NT-proBNP, have a high value in predicting or referring to the diagnosis and severity of acute or chronic HF and are widely used in clinical work (21). However, they still lack some specificity. BNP or NT-proBNP also increases to a certain extent in diseases such as pneumonia, asthma, pulmonary embolism, and interstitial lung disease, and the diagnosis of HF is difficult at present (22). Therefore, the exploration and discovery of more specific biofactors are beneficial to the early screening and diagnosis of HF. Also, determining the initial imbalance and potential mechanism of the disease is urgently needed.
HF is the consequence of the systolic and/or diastolic malfunction of the heart, which causes the venous blood volume to remain in the heart, while the arterial system lacks enough blood perfusion, leading to the buildup of blood in the veins and inadequate blood flow in the arteries (23), resulting in heart and circulatory disease. Immune activation, inflammation, oxidative stress, and mitochondrial biology changes are considered to be important pathophysiological events in this process. The immune system consists of a variety of cellular components, such as granulocytes, mast cells, monocytes, macrophages, dendritic cells, and natural killer (NK) cells (24). A wide range of inflammatory reactions is triggered in the body when damaging stimuli, infections, or tissue damage are present. T and B lymphocytes, which form part of the adaptive immune system, have been found to improve gene coding in HF models. These cytokines can directly promote the development of some pathological states, such as HF, cardiorenal syndrome, pulmonary fibrosis, and cirrhosis. The inflammatory response in HF is closely related to the activation of the immune system. We applied new scientific methods, such as the Boruta algorithm and LASSO logistic regression algorithm, to test the diagnostic factors of HF and analyze the leakage of immune cells in HF (25). However, this study has certain limitations that should be noted. Due to the limited genetic data available, the results may depart from heterotypic cell interactions, disease-induced disorders, or phenotypic plasticity, among other things (26). We also re-examined previously available data sets for the second time as part of our research.
Conclusions
In this study, a total number of 240 transcriptomic data sets were analyzed. We found that SERPINA3 (AUC =0.958) > FCN3 (AUC =0.972) > FREM1 (AUC =0.954) > MNS1 (AUC =0.948) were diagnostic factors of HF. In GSVA, it was shown that FREM1, MNS1, and FCN3 all have a role in bile acid, fatty acid, and heme metabolism, all of which support the theory that central genes have an impact on HF progression through controlling metabolism. In conclusion, SERPINA3-FCN3-MNS1 can be used as a diagnostic factor of HF, and the immune cells in the leakage play an important role in the occurrence and development of HF. Further investigation of these immune cells may help to identify the target of HF immunotherapy and may also aid in the improvement of immunomodulatory treatment for HF patients in the future.
Limitations
Missing experimental data was the biggest short board. So, it was necessary to validate the expressions and its functions of representative genes in HF by real world experimental data.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-22/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-22/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Helsinki Declaration (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat Rev Genet 2004;5:811-25. [Crossref] [PubMed]
- Hannenhalli S, Putt ME, Gilmore JM, et al. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation 2006;114:1269-76. [Crossref] [PubMed]
- Vanburen P, Ma J, Chao S, et al. Blood gene expression signatures associate with heart failure outcomes. Physiol Genomics 2011;43:392-7. [Crossref] [PubMed]
- Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A 2003;100:1896-901. [Crossref] [PubMed]
- Tan FL, Moravec CS, Li J, et al. The gene expression fingerprint of human heart failure. Proc Natl Acad Sci U S A 2002;99:11387-92. [Crossref] [PubMed]
- Margulies KB, Matiwala S, Cornejo C, et al. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res 2005;96:592-9. [Crossref] [PubMed]
- Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008;456:53-9. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- MacDonnell SM, Weisser-Thomas J, Kubo H, et al. CaMKII negatively regulates calcineurin-NFAT signaling in cardiac myocytes. Circ Res 2009;105:316-25. [Crossref] [PubMed]
- Gallagher GL, Jackson CJ, Hunyor SN. Myocardial extracellular matrix remodeling in ischemic heart failure. Front Biosci 2007;12:1410-9. [Crossref] [PubMed]
- Lee HG, Chen Q, Wolfram JA, et al. Cell cycle re-entry and mitochondrial defects in myc-mediated hypertrophic cardiomyopathy and heart failure. PLoS One 2009;4:e7172. [Crossref] [PubMed]
- Miyazaki M, Schroder E, Edelmann SE, et al. Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat. PLoS One 2011;6:e27168. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Yang KC, Yamada KA, Patel AY, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014;129:1009-21. [Crossref] [PubMed]
- Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007;35:W169-75. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Everett LJ, Jensen ST, Hannenhalli S. Transcriptional regulation via TF-modifying enzymes: an integrative model-based analysis. Nucleic Acids Res 2011;39:e78. [Crossref] [PubMed]
- Olson EN, Backs J, McKinsey TA. Control of cardiac hypertrophy and heart failure by histone acetylation/deacetylation. Novartis Found Symp 2006;274:3-12; discussion 13-9, 152-5, 272-6.
- Hu G, Agarwal P. Human disease-drug network based on genomic expression profiles. PLoS One 2009;4:e6536. [Crossref] [PubMed]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118-27. [Crossref] [PubMed]
- Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007;23:3251-3. [Crossref] [PubMed]
- Stafford P, Brun M. Three methods for optimization of crosslaboratory and cross-platform microarray expression data. Nucleic Acids Research 2007;35:e72. [Crossref] [PubMed]
- Chen C, Grennan K, Badner J, et al. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One 2011;6:e17238. [Crossref] [PubMed]
(English Language Editor: A. Kassem)