
A Nomogram model for predicting the occurrence of no-reflow phenomenon after percutaneous coronary intervention using the lncRNA TUG1/miR-30e/NPPB biomarkers
Introduction
Acute myocardial infarction is currently one of the most lethal diseases. The treatment strategy of choice for patients with ST-segment elevation myocardial infarction (STEMI) is direct percutaneous coronary intervention (PCI). However, despite the opening of the offending vessel, the distal myocardium remains ineffectively perfused in some patients. This is referred to as the no-reflow phenomenon, which can lead to swelling and even irreversible damage to the myocardial cells and can thus seriously affect patients’ prognosis (1-3).
A study by Gupta, 2016 reported on the causes and mechanisms of the no-reflow phenomenon after PCI remain inconclusive (4). However, the microvascular embolism/spasm and endothelial injury mechanisms are much related to no-reflow phenomenon (5). Conventional and environmental risk factors such as age, gender, history, smoking, high blood pressure, diabetes, dyslipidaemia are the common risk factor for STEMI (6). The association of dyslipidaemia and diabetes with no-reflow could be due to the spasm and plugging in macrovascular tissues, intravascular thrombus, swelling in endothelial cells, and edema might worsen the intensity of microvascular obstruction leading to both elective and infarct-related PCI (7). C-reactive protein (CRP) is an independent risk predictor in patients with acute coronary disease, and it has been previously shown that PCI has been associated with an early rise in CRP (8).
Recent studies have reported numerous novel and reliable biomarkers have been used in clinical settings (9,10). The novel biomarkers are much needed to integrate within the clinical models to support the clinicians by identifying the patients at high risk for adverse clinical prognosis and provide accurate prognosis estimation with proper prevention program. Regarding to that, we have selected three biomarkers; brain natriuretic peptide precursor B (NPPB), taurine upregulated 1 (TUG1) and micro-ribonucleic acid-30e (miR-30e) that are related to STEMI (11-13).
The NPPB comprises 32 amino acids and is a neurohormone synthesised and secreted by the ventricular muscles as an indicator of ventricular function (14). Studies have shown that polymorphisms in the NPPB gene are strongly associated with diabetes, hypertension, myocardial infarction, and heart failure (11,12). The upstream regulatory pathway of NPPB is currently unclear and needs to be further explored. The overexpression of TUG1 suggests a poor prognosis of heart attack (13). At the onset of STEMI, miR-30e, which is a microRNA (miRNA) associated with cardiomyocyte autophagy and apoptosis, was significantly downregulated. There may be a regulatory relationship between TUG1, miR-30e, and NPPB. Taking this into an account, this study sought to reveal the potential interrelationship between TUG1, miR-30e, and NPPB markers based on clinical relevance combined with bioinformatic methods to determine the mechanism of the no-reflow phenomenon in patients with STEMI after PCI and to improve the diagnosis and prognosis of the patients. However, the exact mechanism still remains unclear and the studies on risk factors affecting reflow are less in number and inconsistent. Hence, more models to determine the relevant risk factors acting no-reflow are much needed to formulate the preventive measures to treat the patients.
Based on the affecting risk factors, a prediction model; Nomogram is a prediction model selected by multi-factor regression analysis could visualize the contribution of each affecting factor to the outcome events with more accuracy and reliability (15). In this study, a nomogram prediction model was established based on the risk factors affecting no-reflow after PCI in patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-481/rc).
Methods
Patient information
A total of 76 patients with STEMI at The Second Affiliated Hospital of Nanchang University were enrolled in the study between 2018 and 2021. They were treated with PCI within 12 hours of the onset of the disease. The diagnostic criteria for STEMI were as follows: clinical symptoms of ischaemic chest pain (>30 minutes), creatine kinase-myocardial isoform, and/or cardiac troponin I values above the upper reference limit, and an electrocardiogram showing new ST-segment elevation or new significant LBBB. Patients were excluded from the study if they met the following exclusion criteria: had myocardial infarction due to non-atherosclerotic factors, such as aortic coarctation and acute non-embolism; had angina pectoris or coronary artery spasm with normal coronary flow; had STEMI to interrupt blood flow after invasive operations; had a severe infection and leukopeania or corticosteroid therapy; had hepatic or renal dysfunction; had cardiogenic shock; had malignant tumours; and/or had other systemic critical diseases. Conventional and the clinical factors were taken from the medical record.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of The Second Affiliated Hospital of Nanchang University (No. 2018-069). Informed consent was obtained from all the patients.
Reflow diagnostics
Coronary flow was comprehensively evaluated before and after PCI according to the thrombolysis in myocardial infarction (TIMI) flow grade (16). The TIMI flow grade classification criteria were as follows: Grade 0, no reperfusion or occlusion, and no flow in distal vascular lesions; Grade 1, partial reperfusion, where the contrast medium penetrated the blockage but did not completely fill the distal coronary artery; Grade 2, partial reperfusion, where the contrast medium filled the entire coronary artery, but the rate of the contrast medium entry and clearance was slower than that of the normal coronary artery; and Grade 3, complete reperfusion, where the contrast medium filled and cleared rapidly in the coronary artery, and the flow rate was consistent with the normal vascular flow rate. Normal reflow was defined as a post-procedural TIMI flow grade of 3 with <20% residual stenosis of the vessels. No-reflow was defined as no significant mechanical obstruction, no significant residual stenosis or entrapment after emergency PCI, but an antegrade flow obstruction (TIMI flow grade ≤2) preventing the effective perfusion of the myocardium with components.
Sample collection
For all the patients, peripheral venous blood was collected within 1 hour of admission before the patient took aspirin, clopidogrel, or heparin, and was analyzed to obtain the complete blood counts and biochemical parameters. These blood samples were collected in tubes containing ethylenediaminetetraacetic acid (BD; Franklin Lakes, NJ, USA) and were centrifuged at 1,900 ×g for 10 minutes at 4 ℃ for 1 hour to remove blood cells. The separated plasma was then centrifuged at 16,000 ×g for 10 minutes at 4 ℃ to remove cellular debris with nucleic acids attached. The centrifuged plasma samples were then transferred to ribonuclease/deoxyribonuclease-free tubes and stored at −80 ℃ for further analysis.
Biochemical parameters
High-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C), high sensitive CRP, serum creatinine and Cystatin C were measured by Beckman Coulter AU5800 automatic biochemical analyzer.
Quantitative real-time PCR (qRT-PCR)
TRIZOL reagent (Invitrogen; Carlsbad, CA, USA) was used to extract total ribonucleic acids (RNAs). For the quantitative analysis of miR-30e, total RNA was isolated using the miRNeasy plasma kit (QIAGEN; Hilden, Germany). According to the manufacturer’s instructions, the total RNA was extracted using stem-loop antisense primer mix and avian myeloblastosis virus transcriptase (TaKaRa; Dalian, China) reverse transcription complementary deoxyribonucleic acid. The SYBR QPCR kit (Toyobo, Osaka, Japan) was then used to perform qRT-PCR on an ABI 7500 real-time PCR system (Applied Biosystems; CA, USA) (17). The primer-pair sequences used were: TUG1, 5'-CTGAAGAAAGGCAACATC-3' (forward) and 5'-GTAGGCTACTACAGGATTTG-3' (reverse); miR-30e, 5'-ACACTCCAGCTGGGT GTAAACATCCTTGAC-3' (forward) and 5'-CTCAACTGGTGTCGTGGAGTCGG CAATTCAGTTGAGCTTCCA-3' (reverse); NPPB, 5'-CTTTCCTGGGAGGTCGT TCC-3' (forward) and 5'-GTTGCGCTGCTCCTGTAAC-3' (reverse); U6, 5'-CTCGCT TCGGCAGCACA-3' (forward) and 5'-AACGCTTCACGAATTTGCGT-3' (reverse); and GAPDH, 5'-AAGAAGGTGGTGAAGCAGGC-3' (forward) and reverse, 5'-TCCACCACCCAGTTGCTGTA-3' (reverse). The quantification of messenger RNA (mRNA) and miRNA was performed using the 2−ΔΔCt method and was normalized by glyceraldehyde-3-phosphate dehydrogenase or U6.
Bioinformatics analysis
Bioinformatics methods were used to predict competitive endogenous RNA (ceRNA) interaction relationships. Interaction sites among long non-coding RNA (lncRNA) TUG1, miR-30e, and NPPB were predicted using RNAhybrid (18) and miRbase (19). The Gene Expression Omnibus (GEO) database was used to retrieve the data of patients with STEMI undergoing PCI. The retrieved GSE61144 transcriptome data were used to analyze the enrichment pathway of lncRNA TUG1. The gene set enrichment analysis (GSEA) of pathways was performed using the cluster Profiler package (20).
Statistical analyses
All the statistical analyses were performed using R software (version 3.6.0). The t-test was used to compare the continuous variables between the 2 groups. The chi-square test or Fisher’s exact test was used to compare discrete variables. The correlation analysis was performed using the Pearson’s test. The receiver operating characteristic (ROC) curves and areas under the curve (AUCs) were used to assess the predictive power of no-reflow occurrences (21). A P value <0.05 was considered statistically significant.
Results
Patient characteristics
Figure 1 illustrates the study flow chart. A total of 76 patients with STEMI were included. Of the patients, 32 had no-reflow and 44 had normal reflow. The basic characteristics of these 2 patient groups are shown in Table 1. Among the 44 patients in the normal-reflow group, there were 33 men and 11 women, with a mean age of 54.89±8.97 years. Among the 32 patients in the no-reflow group, there were 23 men and 9 women, with a mean age of 63.00±11.69 years. Compared to the patients in the normal-reflow group, patients in the no-reflow group were older and a significantly higher percentage had a Killip class >2 at admission, multi-vessel disease, a longer pain-to-balloon time, higher levels of blood glucose, and high-sensitivity C-reactive protein (hs-CRP) levels (P<0.05; Table 1). Conversely, HDL-C and left ventricular ejection fraction (LVEF) at admission were lower in the no-reflow group than the normal-reflow group (P<0.05; Table 1). There were also no significant differences between the 2 groups in terms of gender distribution, history of hypertension, history of diabetes, smoking status, dyslipidemia, body mass index, and other biochemical parameters (P>0.05).
Table 1
| Characteristics | No-reflow (n=32) | Normal reflow (n=44) | t/χ² | P value |
|---|---|---|---|---|
| Age, years | 63.00±11.69 | 54.89±8.97 | 3.285 | 0.002** |
| Male | 23 (71.88) | 33 (75.00) | 0.093 | 0.760 |
| BMI, kg/m2 | 26.71±4.40 | 25.37±3.84 | 1.409 | 0.163 |
| Smoking | 13 (40.63) | 16 (36.36) | 0.143 | 0.706 |
| Hypertension | 14 (43.75) | 14 (31.82) | 1.134 | 0.287 |
| Diabetes mellitus | 17 (53.13) | 14 (31.82) | 3.482 | 0.062 |
| Dyslipidemia | 11 (34.38) | 11 (25.00) | 0.792 | 0.374 |
| Killip class >2 at admission | 6 (18.75) | 3 (6.82) | 2.526 | 0.112 |
| Multi-vessel disease | 23 (71.88) | 19 (43.18) | 6.169 | 0.013* |
| LVEF on admission | 39.69±5.22 | 50.52±6.05 | –8.153 | <0.001** |
| Pain-to-balloon time, min | 244.34±60.31 | 180.45±67.08 | 4.275 | <0.001** |
| WBC, 109/L | 6.33±1.64 | 6.01±1.79 | 0.792 | 0.431 |
| Platelet, 109/L | 252.01±85.52 | 257.02±87.33 | –0.249 | 0.804 |
| Blood glucose, mmol/L | 11.71±1.25 | 10.83±0.84 | 3.467 | 0.001** |
| HDL-C, mmol/L | 1.63±0.40 | 1.83±0.40 | –2.216 | 0.030* |
| LDL-C, mmol/L | 2.77±0.78 | 2.52±0.65 | 1.54 | 0.128 |
| Serum creatinine, μmol/L | 55.32±13.87 | 56.61±13.11 | –0.415 | 0.679 |
| Cystatin C, mg/L | 0.81±0.26 | 0.90±0.26 | –1.533 | 0.129 |
| hs-CRP, mg/L | 25.89±6.53 | 13.03±2.26 | 10.686 | <0.001** |
Data are presented as mean ± standard deviation or number (%). *, P<0.05; **, P<0.01. BMI, body mass index; LVEF, left ventricular ejection fraction; WBC, white blood cell; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; hs-CRP, high-sensitivity C-reactive protein.
Expression levels of lncRNA TUG1/miR-30e/NPPB between the 2 groups
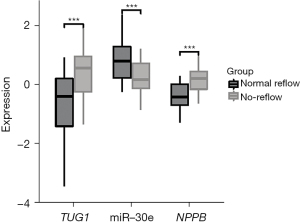
Based on our findings, miR-30e and NPPB expression levels differed between the 2 groups. To investigate the potential role of lncRNA TUG1, miR-30e, and NPPB in no-reflow PCI, we extracted and isolated plasma from patients at the time of admission. We used qRT-PCR to detect TUG1, miR-30e, and NPPB mRNA expression levels in plasma after PCI in 44 and 32 patients with normal reflow and no reflow, respectively. As Figure 2 shows, plasma lncRNA TUG1 and NPPB were more highly expressed in the no-reflow group. In this study, miR-30e was also found to be less expressed in the no-reflow group than the normal-reflow group.
Interaction of plasma TUG1/miR-30e/NPPB axis
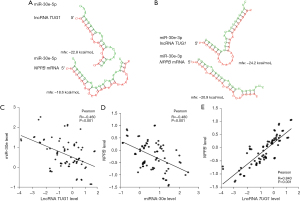
Our bioinformatics analysis predicted that lncRNA TUG1 and miR-30e have multiple binding sites. As Figure 3A,3B show, there are binding sites on the structure of lncRNA TUG1, miR-30e-3p, and miR-30e-5p. MiR-30e-3p and miR-30e-5p were also found to interact with NPPB mRNA. To further validate the interaction between TUG1, miR-30e, and NPPB mRNAs, we performed a Pearson correlation analysis to examine their expression. As Figure 3C shows, we found a negative correlation between lncRNA TUG1 and miR-30e expression in the plasma of patients from the normal-reflow and no-reflow groups. Similarly, there was a negative correlation between miR-30e and NPPB expression (Figure 3D). However, there was a positive correlation between lncRNA TUG1 and NPPB mRNA expression (Figure 3E).
Logistics regression analysis of no-reflow risk factors
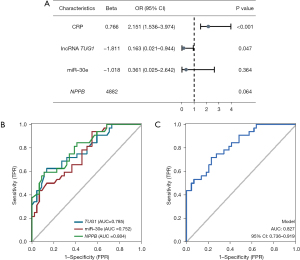
We tested different factors as independent variables in the no-flow logistic regression analysis of risk factor prediction. The univariate analysis identified age, LVEF on admission, pain-to-balloon time, multi-vessel disease, blood glucose, HDL-C, hs-CRP, lncRNA, TUG1, miR-30e, and NPPB as risks (Table 2). Among them, advanced age, low LVEF on admission, long pain-to-balloon time, multi-vessel disease, blood glucose, hs-CRP, lncRNA, TUG1, and NPPB were risk factors for no reflow. Additionally, HDL-C and miR-30e were identified as protective factors for no reflow. We further included hs-CRP, lncRNA, TUG1, miR-30e, and NPPB as independent variables in a multivariate logistic regression analysis, which identified lncRNA TUG1 [odds ratio (OR): 0.163, 95% confidence interval (CI): 0.021–0.944] and hs-CRP (OR: 2.151, 95% CI: 1.536–3.974) as independent predictors (Figure 4A). A model was constructed based on multifactor logistic regression analysis, and the C-index of the model predicting no reflow was obtained as 0.982 (95% CI: 0.956–1.000). This model was validated by internal sampling, and the C-index obtained was 0.975.
Table 2
| Characteristics | OR | 95% CI | P value |
|---|---|---|---|
| Male | 0.852 | 0.304–2.426 | 0.760 |
| Age, years | 1.080 | 1.030–1.139 | 0.002** |
| BMI, kg/m2 | 1.085 | 0.969–1.223 | 0.163 |
| Hypertension | 1.667 | 0.649–4.328 | 0.289 |
| Diabetes mellitus | 2.429 | 0.957–6.337 | 0.064 |
| Dyslipidemia | 1.571 | 0.576–4.314 | 0.375 |
| Smoking | 1.197 | 0.467–3.062 | 0.706 |
| Killip class >2 at admission | 3.154 | 0.763–15.986 | 0.126 |
| LVEF on admission | 0.647 | 0.505–0.768 | <0.001** |
| Pain-to-balloon time, min | 1.015 | 1.007–1.024 | <0.001** |
| Multi-vessel disease | 3.363 | 1.300–9.250 | 0.015* |
| WBC, 109/L | 1.116 | 0.854–1.477 | 0.426 |
| Platelet, 109/L | 0.999 | 0.994–1.005 | 0.801 |
| Blood glucose, mmol/L | 2.343 | 1.433–4.204 | 0.002** |
| HDL-C, mmol/L | 0.273 | 0.077–0.872 | 0.034* |
| LDL-C, mmol/L | 1.683 | 0.873–3.414 | 0.130 |
| Serum creatinine, μmol/L | 0.993 | 0.958–1.027 | 0.674 |
| Cystatin C, mg/L | 0.242 | 0.036–1.460 | 0.131 |
| hs-CRP, mg/L | 1.939 | 1.502–2.917 | <0.001** |
| TUG1 | 3.158 | 1.796–6.340 | <0.001** |
| miR-30e | 0.192 | 0.070–0.447 | <0.001** |
| NPPB | 15.638 | 4.593–69.602 | <0.001** |
*, P<0.05; **, P<0.01. OR, odds ratio; CI, confidence interval; BMI, body mass index; LVEF, left ventricular ejection fraction; WBC, white blood cell; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; hs-CRP, high-sensitivity C-reactive protein; TUG1, taurine upregulated 1; NPPB, natriuretic peptide precursor B.
Predictive value of plasma TUG1/miR-30e/NPPB for no reflow
To investigate whether lncRNA TUG1/miR-30e/NPPB can distinguish between normal reflow and no reflow, we applied logistic regression for the ROC analysis. As Figure 4B shows, the AUCs of TUG1, miRNA-30e, and NPPB in predicting no reflow under a single factor were 0.785, 0.752, and 0.804, respectively. LncRNA TUG1, miRNA-30e, and NPPB were further co-incorporated into the multifactorial analysis. As Figure 4C shows, the predicted AUC of the multifactorial logistic regression model was 0.827 (95% CI: 0.736–0.919). These results suggest that lncRNA TUG1, miRNA-30e, and NPPB can differentiate between normal reflow and no reflow in STEMI after PCI.
GSEA analysis of lncRNA TUG1
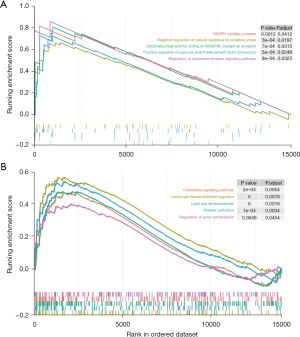
We conducted a GSEA to explore the possible role of the independent predictor lncRNA TUG1. According to the GSEA, the gene ontology pathways associated with TUG1 were the regulation of the dopamine receptor signalling pathway, the regulation of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex, the regulation of oxidoreductase activity, acting on NAD(P)H, oxygen as an acceptor, the positive regulation of vascular endothelial growth factor production, and the negative regulation of cellular response to oxidative stress (Figure 5A). The TUG1-associated Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways were leukocyte trans-endothelial migration, platelet activation, lipid, and atherosclerosis regulation, regulation of the chemokine signalling pathway, and regulation of actin cytoskeleton (Figure 5B). These results suggest that TUG1 is associated with energy metabolism, vascular endothelial growth, oxidative stress, cellular chemotaxis, and atherosclerotic functions. In summary, TUG1 plays an important role in no-reflow PCI among patients with STEMI. We also identified a potential lncRNA TUG1/miR-30e/NPPB pathway that upregulated NPPB in no reflow that may play an important role in the phenomenon.
Discussion
This study aimed to investigate the potential regulatory relationship among TUG1, miR-30e, and NPPB, to screen for no-reflow risk factors through clinical data combined via a bioinformatics analysis, and to develop a nomogram prediction model. The results showed that lncRNA TUG1, miR-30e, NPPB, and hs-CRP were risk factors for patients with STEMI after PCI no reflow. The clinical prediction model based on lncRNA TUG1, miR-30e, NPPB, and hs-CRP also had good predictive power. LncRNA TUG1 and miR-30e expression and miR-30e and NPPB expression were both negatively correlated. Conversely, lncRNA TUG1 and NPPB mRNA expression were positively correlated. We also found that the lncRNA TUG1/miR-30e/NPPB pathway was upregulated during STEMI. Thus, lncRNA TUG1 may be a molecular sponge for miR-30e to promote NPPB expression, ultimately leading to cardiomyocyte injury and the onset of the no-reflow phenomenon after PCI.
LncRNAs regulate genes epigenetically, transcriptionally, post-transcriptionally, transliterally, and post-translationally. TUG1 is a member of the lncRNA family, which regulates gene expression at multiple levels. TUG1 reportedly participates in and mediates various biological processes. A study has shown that lncRNA TUG1 is involved in the progression of multiple cardiovascular-related diseases (22). In the cardiovascular context, TUG1 promotes hypoxia-induced injury and induces cardiomyocyte death (23). In the present study, we found that TUG1 was associated with energy metabolism, vascular endothelial growth, oxidative stress, cellular chemotaxis, and atherosclerosis. LncRNA TUG1 expression was also negatively correlated with miR-30e and was positively correlated with that of NPPB mRNA.
MiRNAs are important regulators of cardiac homeostasis and stress responses. By disrupting mRNAs that encode proteins in cellular metabolism, growth, calcium signalling, and programmed death pathways, microRNAs exert control over key biological processes, and are important regulators of the stable internal environment of cardiomyocytes (24,25). It has been shown that miR-30e is associated with cardiomyocyte protection after myocardial ischaemia/reperfusion (26). Members of the miR-30 family are abundantly expressed in cardiac tissue and are extensively involved in the pathophysiology of a variety of heart diseases (27-29). The miR-30 family also plays a protective role in diseases, such as myocardial hypertrophy, myocardial infarction, heart failure, and ischaemia-reperfusion injury (30-33). Further, a previous study showed that miR-30e-3p expression was significantly reduced after coronary microembolization (CME) (34). Autophagy is a mechanism that maintains homeostasis in cardiomyocytes (35). MiR-30e-3p has also been found to be involved in CME-induced cardiac dysfunction by possibly regulating myocardial autophagy (34). In the present study, miR-30e expression was found to be lower in the no-reflow group than the normal-reflow group. The bioinformatics analysis also predicted multiple binding sites for lncRNA TUG1 and miR-30e and found that miR-30e-3p and miR-30e-5p interact with NPPB mRNA.
Hs-CRP, a cytokine closely associated with inflammatory responses, is reportedly strongly associated with the development of cardiovascular disease and is recognised as a strong predictor of cardiovascular risk events (36,37). In the univariate logistic regression analysis of the present study, hs-CRP, lncRNA, TUG1, miRNA-30e, and NPPB were identified as risk variables. Further, lncRNA TUG1 (OR: 0.163; 95% CI: 0.021–0.944) and hs-CRP (OR: 2.151; 95% CI: 1.536–3.974) were identified as independent predictors. However, the present study had a limited number of clinical samples, future prospective studies are needed with large samples, multi-regional, multicentre and multigene studies to verify the reliability of the nomogram's predictive ability.
Conclusions
This study was the first to reveal the potential regulatory relationship among TUG1, miR-30e, and NPPB. Clinical prediction models based on lncRNA TUG1, miR-30e, NPPB, and hs-CRP have good predictive power based on clinical data combined via a bioinformatic analysis. The potential mechanism of the no-reflow phenomenon in patients with STEMI after PCI was described, which provides new approaches and new therapeutic targets for the clinical management of STEMI.
Acknowledgments
Funding: This work was supported by the Key Research and Development Program of Guangxi (grant No. AB20159005), and National College Students Innovation and Entrepreneurship Training Program (grant No. 202110601001).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-481/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-481/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-481/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of The Second Affiliated Hospital of Nanchang University (No. 2018-069). Informed consent was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang Z, Ren L, Liu N, et al. Utility of Hematological Parameters in Predicting No-Reflow Phenomenon After Primary Percutaneous Coronary Intervention in Patients With ST-Segment Elevation Myocardial Infarction. Clin Appl Thromb Hemost 2018;24:1177-83. [Crossref] [PubMed]
- Alizadehsani R, Hosseini MJ, Boghrati R, et al. Exerting Cost-Sensitive and Feature Creation Algorithms for Coronary Artery Disease Diagnosis. International Journal of Knowledge Discovery in Bioinformatics 2012;3:59-79. [Crossref]
- Sensoy B, Uzunget SB, Acikgoz S, et al. Renal Dysfunction on Admission Predicts No-Reflow Phenomenon in Patients Undergoing Manual Thrombus Aspiration during Primary Percutaneous Coronary Intervention. Acta Cardiol Sin 2016;32:185-93. [PubMed]
- Gupta S, Gupta MM. No reflow phenomenon in percutaneous coronary interventions in ST-segment elevation myocardial infarction. Indian Heart J 2016;68:539-51. [Crossref] [PubMed]
- Pan HF. Progress of no-reflow in patients with acute myocardial infarction undergoing percutaneous coronary intervention. Adv Cardiovasc Dis 2018;39:1029-34.
- Yunyun W, Tong L, Yingwu L, et al. Analysis of risk factors of ST-segment elevation myocardial infarction in young patients. BMC Cardiovasc Disord 2014;14:179. [Crossref] [PubMed]
- Jaffe R, Charron T, Puley G, et al. Microvascular obstruction and the no-reflow phenomenon after percutaneous coronary intervention. Circulation 2008;117:3152-6. [Crossref] [PubMed]
- Ray KK, Nazer B, Cairns R, et al. Association between percutaneous coronary intervention and long-term C-reactive protein levels in patients with acute coronary syndromes. J Thromb Thrombolysis 2010;30:10-3. [Crossref] [PubMed]
- Zhao E, Xie H, Zhang Y. A Nomogram Based on Apelin-12 for the Prediction of Major Adverse Cardiovascular Events after Percutaneous Coronary Intervention among Patients with ST-Segment Elevation Myocardial Infarction. Cardiovasc Ther 2020;2020:9416803. [Crossref] [PubMed]
- Yang L, Cong H, Lu Y, et al. A nomogram for predicting the risk of no-reflow after primary percutaneous coronary intervention in elderly patients with ST-segment elevation myocardial infarction. Ann Transl Med 2021;9:126. [Crossref] [PubMed]
- Pereira NL, Tosakulwong N, Scott CG, et al. Circulating atrial natriuretic peptide genetic association study identifies a novel gene cluster associated with stroke in whites. Circ Cardiovasc Genet 2015;8:141-9. [Crossref] [PubMed]
- Fedorowski A, Franceschini N, Brody J, et al. Orthostatic hypotension and novel blood pressure-associated gene variants: Genetics of Postural Hemodynamics (GPH) Consortium. Eur Heart J 2012;33:2331-41. [Crossref] [PubMed]
- Li B, Wu Y. LncRNA TUG1 overexpression promotes apoptosis of cardiomyocytes and predicts poor prognosis of myocardial infarction. J Clin Pharm Ther 2020;45:1452-6. [Crossref] [PubMed]
- Eindhoven JA, van den Bosch AE, Jansen PR, et al. The usefulness of brain natriuretic peptide in complex congenital heart disease: a systematic review. J Am Coll Cardiol 2012;60:2140-9. [Crossref] [PubMed]
- Zhang Z, Ji Z, Du R, et al. Establishment and Evaluation of Nomogram Model for Predicting the Risk of No Reflow after Percutaneous Coronary Intervention in Patients with Acute Myocardial Infarction. Indian J Pharm Sci 2021;30:217-23.
- Ganz W. The thrombolysis in myocardial infarction (TIMI) trial. N Engl J Med 1985;313:1018. [Crossref] [PubMed]
- Lin J, Luo Z, Liu S, et al. Long non-coding RNA H19 promotes myoblast fibrogenesis via regulating the miR-20a-5p-Tgfbr2 axis. Clin Exp Pharmacol Physiol 2021;48:921-31. [Crossref] [PubMed]
- Rehmsmeier M, Steffen P, Hochsmann M, et al. Fast and effective prediction of microRNA/target duplexes. RNA 2004;10:1507-17. [Crossref] [PubMed]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155-62. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Xu Y, Wang K, Wang Q, et al. The Antioxidant Enzyme PON1: A Potential Prognostic Predictor of Acute Ischemic Stroke. Oxid Med Cell Longev 2021;2021:6677111. [Crossref] [PubMed]
- Li FP, Lin DQ, Gao LY. LncRNA TUG1 promotes proliferation of vascular smooth muscle cell and atherosclerosis through regulating miRNA-21/PTEN axis. Eur Rev Med Pharmacol Sci 2018;22:7439-47. [PubMed]
- Wu Z, Zhao S, Li C, et al. LncRNA TUG1 serves an important role in hypoxia-induced myocardial cell injury by regulating the miR-145-5p-Binp3 axis. Mol Med Rep 2018;17:2422-30. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Dorn GW 2nd. MicroRNAs: redefining mechanisms in cardiac disease. J Cardiovasc Pharmacol 2010;56:589-95. [Crossref] [PubMed]
- Baars T, Skyschally A, Klein-Hitpass L, et al. microRNA expression and its potential role in cardioprotection by ischemic postconditioning in pigs. Pflugers Arch 2014;466:1953-61. [Crossref] [PubMed]
- Li J, Donath S, Li Y, et al. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet 2010;6:e1000795. [Crossref] [PubMed]
- Zhang T, Tian F, Wang J, et al. Endothelial Cell Autophagy in Atherosclerosis is Regulated by miR-30-Mediated Translational Control of ATG6. Cell Physiol Biochem 2015;37:1369-78. [Crossref] [PubMed]
- Chen M, Ma G, Yue Y, et al. Downregulation of the miR-30 family microRNAs contributes to endoplasmic reticulum stress in cardiac muscle and vascular smooth muscle cells. Int J Cardiol 2014;173:65-73. [Crossref] [PubMed]
- Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 2009;104:170-8, 6p following 178.
- Piccolo R, Oliva A, Avvedimento M, et al. Mortality after bleeding versus myocardial infarction in coronary artery disease: a systematic review and meta-analysis. EuroIntervention 2021;17:550-60. [Crossref] [PubMed]
- Yu YY, Zhao BW, Ma L, et al. Association Between Out-of-Hour Admission and Short- and Long-Term Mortality in Acute Myocardial Infarction: A Systematic Review and Meta-Analysis. Front Cardiovasc Med 2021;8:752675. [Crossref] [PubMed]
- Shen Y, Shen Z, Miao L, et al. miRNA-30 family inhibition protects against cardiac ischemic injury by regulating cystathionine-γ-lyase expression. Antioxid Redox Signal 2015;22:224-40. [Crossref] [PubMed]
- Wang XT, Wu XD, Lu YX, et al. Potential Involvement of MiR-30e-3p in Myocardial Injury Induced by Coronary Microembolization via Autophagy Activation. Cell Physiol Biochem 2017;44:1995-2004. [Crossref] [PubMed]
- Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 2007;13:619-24. [Crossref] [PubMed]
- Liu HH, Cao YX, Sun D, et al. High-sensitivity C-reactive protein and hypertension: combined effects on coronary severity and cardiovascular outcomes. Hypertens Res 2019;42:1783-93. [Crossref] [PubMed]
- Gregory MA, Manuel-Apolinar L, Sánchez-Garcia S, et al. Soluble Intercellular Adhesion Molecule-1 (sICAM-1) as a Biomarker of Vascular Cognitive Impairment in Older Adults. Dement Geriatr Cogn Disord 2019;47:243-53. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


