
Research progress regarding long-chain non-coding RNA in lung cancer: a narrative review
Introduction
Background
At the pathological level, lung cancer is classified into small-cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), and is the main cause of cancer-related death worldwide, with a high incidence (1). About 80% of primary lung cancers are NSCLCs, which can be further divided into lung adenocarcinoma (LAD), large cell carcinoma (LCC), and lung squamous cell carcinoma (LSCC) (2,3). A significant number of patients miss the ideal opportunity for surgical treatment as they are first diagnosed in the advanced stage, which is partly attributable to the lack of biomarkers for early diagnosis (4).
Previous studies have reported that long non-coding ribonucleic acids (lncRNAs) are involved in regulating the transcription and translation of protein-coding genes and play an important role in tumor growth, metastasis, and drug resistance (5,6). In this paper, the abnormally expressed lncRNAs in lung cancer were summarized, and the mechanism of regulating tumor drug resistance and radiotherapy sensitivity was discussed in depth. This paper provides new insights for the early diagnosis and targeted therapy of lung cancer, and also proposes a basis for identifying biomarkers that predict tumor sensitivity to chemotherapy and radiotherapy.
Overview of the lung cancer screening methods
At present, the methods of lung cancer screening mainly include X-ray, low-dose computed tomography (LDCT), positron emission tomography-computed tomography (PET/CT), electronic bronchoscopy, and serum tumor marker detection. X-ray chest film is used to screen lung cancers; however, its sensitivity and detection rate are extremely low. LDCT increases the detection rate of lung cancer to a certain extent. PET/CT has high specificity and sensitivity, especially in the diagnosis, staging, and curative effect evaluation of lung cancer. Electronic bronchoscopy is applied for the early diagnosis of lung cancer which is an invasive operation. The detection of serum tumor markers, such as carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), cytokeratin 19 fragments (CYFRA 21-1), and epidermal growth factor receptor (EGFR), are important auxiliary means for the diagnosis and evaluation of the pathological types and clinical stages of lung cancer (7). However, there are still clinical challenges in improving the patient survival rate due unsatisfied early diagnosis based on the traditional methods (8,9). Thus, biomarkers for early diagnosis and prognosis of lung cancer are extremely needed. LncRNAs have been in the stage of research for numerous unknown functions while the available clinical application potential is explicit for the exact evidences in clinical researches. Many recent clinical studies have shown that it has caused widespread attention for its characteristics including high specificity of tissue, high serological stability in body fluids, easily accessible detection in serum exosomes as well as the high accuracy of signature diagnosis and prediction (10-13). Tumor-specific lncRNA are potential markers for early diagnosis and treatment of cancer. A previous study reported that a variety of lncRNAs were expressed at abnormal levels in patients with lung cancer, some of which are related to tumor drug resistance (14). This indicates that lncRNA may be used as a potential biomarker for the diagnosis and prognostic evaluation of lung cancer.
Biological functions of lncRNA
LncRNAs are a kind of noncoding RNAs (ncRNA) with a length >200 nucleotides which are currently the primary and most concerned ncRNAs together with the microRNA (miRNA) and circle RNA (circRNA) in cancer research. Report has showed that lncRNAs may be involved in more diverse and complex mechanisms to regulating more biogenetic process because of the longer sequences and more complex spatial structures compared with small RNAs (15). Thus, there are more potential lncRNAs to be found as the biomarkers of specific diseases including lung cancer based on a better understand of their vast and complex roles in their pathological regulation.
LncRNAs participate in numerous physiological and pathological processes by interacting with deoxyribonucleic acid (DNA), RNA, and protein at the transcriptional, translational, and epigenetic levels (16), and play a vital role in the proliferation, metastasis, and drug resistance of lung cancer (17). At the transcriptional level, the molecular mechanism of lncRNA participation is as follows: (I) lncRNAs can combine with a gene promoter and regulate the transcription of downstream genes; and (II) lncRNAs lead chromatin-modifying enzymes to specific genomic locus target genes. At the translational level, the molecular mechanism of lncRNA participation is as follows: (I) lncRNAs can be processed into miRNA, and act as the transcriptional precursor of miRNA (18); (II) affecting the stability of messenger RNA (mRNA) by binding to its target mRNA; (III) regulating mRNA splicing patterns and producing different splicing variants; and (IV) the interaction between lncRNA and miRNA: lncRNA with miRNA binding site can degrade miRNA targeted (19). At the epigenetic level, the molecular mechanism of lncRNA participation is as follows: (I) lncRNA can recruit a variety of proteins to form (20); (II) lncRNA regulates histone activity through acetylation, methylation, and ubiquitination (21); and (III) lncRNA is directly related to signaling mediators, such as receptors, protein kinases, and transcription factors, and regulates their enzyme activities (22). Researches have shown that the expression level of lncRNA is significantly different between cancerous and normal tissues, and plays an important role in the occurrence, development, and drug resistance of lung cancer. We present the following article in accordance with the Narrative Review reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-897/rc).
Methods
The narrative review was performed via searching all related scientific reports published from 09/16/2011 to 05/02/2022 in PubMed database. “lncRNA AND lung cancer”, “lncRNA AND non-small-cell lung cancer”, “lncRNA AND drug resistance”, “lncRNA AND radio sensitivity” were used as the key search terms. Research articles with outcomes available to readers written in English were considered. Two authors conducted the full text of the relevant and sophisticated literatures to reach consensus and to discuss among the research members if necessary (Table 1).
Table 1
| Items | Specification |
|---|---|
| Date of Search (specified to date, month and year) | 12/31/2021–5/31/2022 |
| Databases and other sources searched | PubMed |
| Search terms used (including MeSH and free text search terms and filters) | “lncRNA AND lung cancer”, “lncRNA AND non-small-cell lung cancer”, “lncRNA AND drug resistance”, “lncRNA AND radio sensitivity” |
| Timeframe | 09/16/2011–05/02/2022 |
| Inclusion and exclusion criteria (study type, language restrictions etc.) | Research articles with outcomes available to readers written in English were considered |
| Selection process (who conducted the selection, whether it was conducted independently, how consensus was obtained, etc.) | Two authors conducted the full text of the relevant and sophisticated literatures to reach consensus and to discuss among the research members if necessary. |
| Any additional considerations, if applicable | N/A |
Discussion
Abnormal expression of lncRNAs in lung cancer
LncRNAs participate in the occurrence and development of lung cancer, regulate basic cytological processes such as proliferation, cell growth, apoptosis, migration, stem cell maintenance, and epithelial-mesenchymal transition (EMT), and can also serve as signal transduction molecules, molecular decoy, and scaffolds (22-25). Abnormal expression of lncRNAs in lung cancer were summarized below.
Up-regulated lncRNAs in lung cancer
Smoking and cancer-related lncRNA1 (lncRNA-SCAL1)
LncRNA-SCAL1, which is located at chromosome 5q14.3, is related to smoking and cancer. Cigarette smoke extract can induce high expression of lncRNA-SCAL1 in lung cancer cells. Thai et al. found that lncRNA-SCAL1 was increased in the airway epithelial cells and lung cancer cell lines of smokers (26). Overexpression of lncRNA-SCAL1 in lung cells treated with high glucose can inhibit the expression of the inducible nitric oxide synthase (iNOS) protein and reduce the production of nitric oxide (27). These findings indicate that lncRNA-SCAL1 plays an important role in the antioxidant pathway.
Metastasis-associated lung adenocarcinoma transcript 1 (lncRNA-MALAT1)
LncRNA-MALAT1was one of the first lncRNAs to be identified as related to lung cancer. It is also known as nuclear-enriched autosomal transcript 2 (NEAT2), which is a highly conserved nuclear lncRNA. LncRNA-MALAT1 promotes the spread and invasion of lung cancer by regulating the expressions of miR-124/STAT3 and miR-206/AKT (28,29). It has been reported in the literature that the MALAT1 protein level is up-regulated in the tumor tissues of NSCLC patients compared with adjacent tissues (30). Also, compared with the healthy control group, the expression of lncRNA-MALAT1 in the peripheral blood of NSCLC patients is higher and has higher specificity and sensitivity (area under the ROC curve, AUC =0.79) (31). In addition, the overexpression of lncRNA-MALAT1 in the serum exosomes of NSCLC patients was positively correlated with the tumor stage and lymph node metastasis (32). Moreover, knockdown of lncRNA-MALAT1 suppressed the progression of NSCLC by inhibiting growth and metastasis and facilitating apoptosis, possibly by upregulating miR-185-5p and decreasing the expression of MDM4 in NSCLC (33). LncRNA-MALAT1 can be used as a biomarker for the diagnosis and prognosis of NSCLC.
Antisense noncoding RNA in the INK4 Locus (lncRNA-ANRIL)
LncRNA-ANRIL is located on chromosome 9p21.3. The lncRNA ANRIL is located in the nucleus and directly binds to the enhancer of Zeste homolog 2 (EZH2) to increase the binding of EZH2 to Krüppel-like factor 2 (KLF2) and the p21 promoter (34). This promotes the proliferation of NSCLC cells and inhibits apoptosis. Studies have shown that the knockdown of lncRNA-ANRIL can induce cell cycle arrest in the G1/G0 phase and promote apoptosis. The depletion of lncRNA-ANRIL increases p15 expression and induces stagnation of the cell cycle of lung cancer in the G2/M phase (35). Also, compared with healthy people, the expression of lncRNA-ANRIL in tumor tissues and serum samples of NSCLC patients is significantly increased (36,37). LncRNA-ANRIL is overexpressed in NSCLC tissues and cell lines, and the increased expression level of lncRNA-ANRIL is related to the poor prognosis of NSCLC patients (38). LncRNA-ANRIL also increases markedly in the plasma samples of NSCLC patients, with an AUC of 0.798, suggesting that lncRNA-ANRIL in external circulation can be used as a sensitive diagnostic tool (39).
LncRNA-H19
The lncRNA-H19 is a 2.3 kb RNA encoded by the H19 gene and is highly expressed in lung cancer tissues and cells. According to previous literature reports, lncRNA-H19 can be attached to miR-17 as a competitive endogenous RNA (ceRNA) in lung cancer to regulate the expression of signal transducer and activator of transcription (STAT3) (40). It regulates the expression of ROCK2 by combining with miR-484 to activate the ROCK2/JNK pathway (41), thereby promoting the development of lung cancer. In addition, miR-196b directly targets LIN28B (a conserved RNA binding protein) to inhibit LIN28B expression. LncRNA-H19 can adsorb miR-196b by sponge, which reduces the inhibition of miR-196b on LIN28B and increases the expression of LIN28B, thus accelerating the proliferation of lung cancer cells (42). Study has also shown that the expression level of lncRNA-H19 in NSCLC is correlated with tumor size, invasion, and metastasis (43).
Long-chain non-coding growth arrest-specific protein 6-antisense RNA1 (lncRNA-DLX6-AS1)
The lncRNA-DLX6-AS1 is located on chromosome 7q21.3. Its expression level in LAD is up-regulated compared with that in normal adjacent tissues. LncRNA-DLX6-AS1 is involved in regulating the miR-27b-3p/GSPT1 axis to promote the proliferation of lung cancer cells (44). The expression of lncRNA-DLX6-AS1 is up-regulated in NSCLC patients, which is related to the differentiation degree and tumor-node-metastasis (TNM) stage of lung cancer. In NSCLC patients, up-regulation of lncRNA-DLX6-AS1 can accelerate cell proliferation and inhibit apoptosis (45), which suggests that lncRNA-DLX6-AS1 may become a new molecular marker and a potential target for anti-tumor drugs in lung cancer diagnosis.
Plasmacytoma variant translocation 1 (lncRNA-PVT1)
The lncRNA-PVT1 is located in the 8q24 region and on the sense strand of the chromosome. It has been demonstrated that lncRNA-PVT1 can regulate the expression of miR-497 and competitively bind to miR-200a and miR-200b, increase the expression of matrix metalloproteinase 9 (MMP9), to promote the metastasis of NSCLC (46,47). Moreover, it has been reported that lncRNA-PVT1 is overexpressed in NSCLC and the increased lncRNA-PVT1 expression level is closely related to poor prognosis (48). LncRNA-PVT1 is also over-expressed and positively correlates with the clinical stage, lymph node metastasis, and distant metastasis of SCLC patients. Multivariate analysis has shown that lncRNA-PVT1 over-expression may be an independent factor of poor prognosis (49) despite the needed reports for NSCLC. Elevated levels of lncRNA-PVT1 promote lung cancer cell proliferation and metastasis both in vitro and in vivo. LncRNA-PVT1 competes endogenously with miR-128 in the regulation of vascular endothelial growth factor C (VEGFC) expression, which is significantly associated with an unfavorable prognosis in lung cancer (50). In addition, lncRNA-PVT1 targets the miRNA-526b/EZH2 regulatory loop to promote the development of NSCLC when knockdown of lncRNA-PVT1 significantly weakens the proliferation and migration ability of cells (51). These findings indicate that the lncRNA-PVT1 may be a carcinogenic lncRNA, but it can be used as a potential target for treating lung cancer and a biomarker for the prognostic evaluation of lung cancer.
HOX transcript antisense RNA (lncRNA-HOTAIR)
Reversely transcribed from 12q13 human HOXC genes, lncRNA-HOTAIR is the first lncRNA reported to be associated with malignant tumors (52). Clinicopathological correlation analysis shows that the upregulation of lncRNA-HOTAIR is closely associated with lymphatic metastasis and TNM staging. Moreover, the exosome can promote NSCLC proliferation and migration through lncRNA-HOTAIR transportation (53). Exosomal lncRNA-HOTAIR promotes lung cancer cell progression by sponging miR-203 (54). Overexpression of lncRNA-HOTAIR promotes the migration and invasion abilities of lung cancer cells, which are suppressed by the overexpression of miR-149-5p (55).
Down-regulated lncRNA in lung cancer
Growth arrest-specific transcript 5 (lncRNA-GAS5)
LncRNA-GAS5 is a lncRNA that is related to cell proliferation and is located on human chromosome 1q25.1. Insulin-like growth factor 1 (IGF-1) is involved in regulating the proliferation, migration, and apoptosis of cancer cells, and inhibiting IGF-1 can block tumor growth (56). LncRNA-GAS5 can regulate the survival rate of lung cancer cells by regulating the expression of IGF-1R and IGF-1 (57). Research has shown that lncRNA-GAS5 is a tumor suppressor and its expression level in NSCLC is low. Compared with healthy individuals, the expression of lncRNA-GAS5 in lung cancer tissues and plasma of NSCLC patients is significantly decreased (58,59). The xenotransplantation model experiment confirmed that lncRNA-GAS5 overexpression can increase the radiosensitivity of NSCLC cells in vivo and inhibit the occurrence and development of tumors by inhibiting the proliferation and invasion of tumor cells and inducing their apoptosis (60). lncRNA-GAS5 blocks the progression of NSCLC partly by increasing the IRF2 expression level via repression of miR-221-3p (61). In addition, lncRNA-GAS5 can influence the invasion and metastasis of lung cancer through the EMT process (62). These findings shed light on the prospect of lncRNA-GAS5 as a therapeutic target for lung cancer. The above studies suggest that lncRNA-GAS5 is expected to be a biomarker for the diagnosis or prognosis of lung cancer.
Maternal expression gene 3 (MEG3)
LncRNA-MEG3, located at 14q32.2 of the human chromosome, is a kind of cancer-inhibiting lncRNA. P53 is an important transcription factor, which can regulate the expression of multiple target genes and plays a role in inhibiting the development of various cancers, including lung cancer (63). LncRNA-MEG3 can activate p53 to arrest the cell cycle of NSCLC and promote apoptosis (64). It has been reported that MEG3 overexpression induces increased p53 protein expression, which can reduce the proliferation of NSCLC cells in vitro and hinder tumorigenesis in vivo (65). Through real-time fluorescence quantitative polymerase chain reaction (qRT-PCR), it was found that the expression of lncRNA-MEG3 in NSCLC tissues and A549 and HCC823 cell lines were significantly lower than those in the normal group. LncRNA MEG3 can regulate the expression of BRCA1 through competitively binding to microRNA-7-5p (66). LncRNA MEG3 inhibited cell proliferation, migration, invasion and telomerase activity by downregulating DKC1 (67). In addition, lncRNA-MEG3 is lowly expressed in NSCLC and affects the immunity and autophagy of NSCLC cells by regulating the miR-543/IDO signaling pathway (68). The above studies indicate that lncRNA-MEG3 is involved in regulating the occurrence and development of SCLC, and can be used as a potential molecular marker for evaluating the prognosis of SCLC.
BRAF-activated non-protein coding RNA (lncRNA-BANCR)
LncRNA-BANCR is a 693 bp anti-tumor lncRNA located on chromosome 9q21.11. lncRNA-BANCR can inhibit the activation of p38 mitogen-activated protein kinase (MAPK) and JNK, thereby inhibiting the proliferation and migration of lung cancer cells (69). The expression level of lncRNA-BANCR in 30 NSCLC tissues and cell lines were detected by qPCR. The results showed that the expression of lncRNA-BANCR in NSCLC tissues and cells was significantly down-regulated compared with normal lung tissues and cells (70). Furthermore, other studies have shown that lncRNA-BANCR can inhibit the proliferation and invasion of NSCLC cells by regulating the expression level of EMT markers E-cadherin and Vimentin (71). In 113 NSCLC tissue samples, 89 cases had down-regulated BANCR expression, and the expressed level of lncRNA-BANCR in NSCLC patients with shorter survival times decreased significantly (72), suggesting that lncRNA-BANCR can be used as a biomarker to evaluate the prognosis of NSCLC patients.
MIR22 host gene (lncRNA-MIR22HG)
LncRNA-MIR22HG can reportedly be used as a prognostic indicator of hepatocellular carcinoma (73). LncRNA-MIR22HG can inhibit cancer in lung cancer. The expression of Y-box binding protein 1 (YBX1), MET, and p21 can be regulated by inhibiting the expression of lncRNA-MIR22HG, thereby regulating the cell survival and apoptosis signaling pathways (74). RNA sequencing has been performed previously on lung cancer, normal lung tissues, and lung cancer cell lines, and the expression profile of lncRNAs in the data was comprehensively analyzed. The results showed that the expression level of lncRNA-MIR22HG in lung cancer was significantly down-regulated and the low expression of lncRNA-MIR22HG was positively correlated with the low survival rate of patients (75), suggesting that lncRNA-MIR22HG has potential as a new diagnostic/prognostic marker and therapeutic target for lung cancer.
P53-induced cancer-related RNA transcript 1 (lncRNA-PICART1)
The expression of lncRNA-PICART1 is down-regulated in human lung cancer tissues and cell lines, and the knockdown of lncRNA-PICART1 can increase the cell viability of lung cancer cell lines. Overexpression of PICART1 suppressed cell growth, cell colony formation and cell invasion partly though regulating the AKT signaling pathway in NSCLC (76). Furthermore, overexpression of lncRNA-PICART1 also promotes the up-regulation of e-cadherin and the down-regulation of Twist1, MMP2, and MMP9, thus inhibiting the migration of tumor cells. In addition, lncRNA-PICART1 can inhibit the proliferation and promote the apoptosis of lung cancer cells by inhibiting the JAK2/STAT3 signaling pathway (77).
Promoter of CDKN1A antisense DNA damages activated RNA (lncRNA-PANDAR)
LncRNA-PANDAR is located on chromosome 6q21.2, and its expression level in NSCLC cancer tissues is down-regulated compared with adjacent normal tissues. The expression of lncRNA-PANDAR is negatively correlated with tumor size and TNM stage (78). Meanwhile, the high expression of PANDAR increased BECN1 expression levels. Another study showed that the low expression of lncRNA-PANDAR increases the binding between NF-YA and the Bcl-2 promoter, thereby inhibiting the apoptosis of NSCLC cells. Further experiments have shown that the low expression of lncRNA-PANDAR predicts poor prognosis in NSCLC patients (79).
The related lncRNAs as well as their mechanisms of action we summarized and analyzed were shown in Table 2.
Table 2
| LncRNAs | Expression | Regulatory mechanism | Effect | References |
|---|---|---|---|---|
| SCAL1 | Upregulated | Not available | Anti-oxidation | (26,27) |
| MALAT1 | Upregulated | miR-124/STAT3 miR-206/AKT miR-185-5p/MDM4 |
Facilitate diffusion and invasion, proliferation, and migration | (28–33) |
| ANRIL | Upregulated | EZH2 p15 |
Promote proliferation and inhibit apoptosis | (34–39) |
| H19 | Upregulated | miR-17/STAT3 miR-484/ROCK2/JNK miR-196b/LIN28B |
Promote proliferation, migration, invasion, EMT | (40–43) |
| DLX6-AS1 | Upregulated | miR-27b-3p/GSPT1 | Promote proliferation and inhibit apoptosis | (44,45) |
| miR-144/PRR11 | ||||
| PVT1 | Upregulated | miR-497 miR-200a(b)/MMP9 miR-128/VEGFC miRNA-526b/EZH2 |
Promote proliferation, invasion and inhibit apoptosis | (46–48,50,51) |
| HOTAIR | Upregulated | miR-203 miR-149-5p |
Promote progression | (53–55) |
| GAS5 | Downregulated | IGF-1R/IGF-1 miR-221-3p/IRF2 |
Inhibit proliferation, invasion, migration, EMT and induce apoptosis | (57–62) |
| MEG3 | Downregulated | P53 miR-7-5p/ BRCA1 miR-543/IDO |
Inhibit proliferation and induce apoptosis | (65-68) |
| BANCR | Downregulated | MAPK/JNK | Inhibition of proliferation, migration, and invasion | (69–72) |
| MIR22HG | Downregulated | YBX1, MET, p21 | Suppress tumor | (74,75) |
| PICART1 | Downregulated | AKT1 JAK2/STAT3 |
Suppress cell growth, invasion proliferation and induce apoptosis | (76,77) |
| PANDAR | Downregulated | BECN1 NF-YA/Bcl-2 |
Inhibit proliferation, promote apoptosis | (78,79) |
LncRNA and drug resistance
The treatment drugs for lung cancer mainly include cisplatin chemotherapy and molecular targeted drugs, such as epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) (80). However, anti-tumor drug resistance is the main factor leading to treatment failure (81). At present, it has been confirmed that the imbalance of lncRNA is related to lymph node metastasis and poor prognosis of patients, and plays an important role in the drug resistance mechanism of many chemotherapy drugs (82,83). The mechanism of lncRNAs related to drug resistance shown in Figure 1.
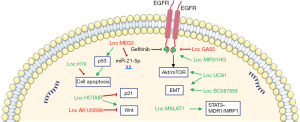
LncRNA and cisplatin resistance
Cisplatin (cis-Diamminedichloroplatinum, CDDP) is a chemotherapeutic drug used in the treatment of lung cancer, which acts on DNA to form a DDP-DNA complex, thus interfering with DNA replication. Cisplatin resistance is the main cause of chemotherapy failure (84). Therefore, studying the role of lncRNAs in cisplatin resistance is crucial to improving the efficacy of chemotherapy in lung cancer patients.
LncRNA-H19-mediated cell proliferation inhibition, and cancer cell metastasis is related to G0/G1 cell cycle arrest, and increased apoptosis (85). Overexpression of the lncRNA-H19 is negatively correlated with cisplatin-based chemotherapy in patients (86). The lncRNA-MALAT1 upregulates MRP1 and MDR1 via STAT3 activation, thus increasing the cisplatin resistance of lung cancer (87). Also, overexpressed level of lncRNA-HOTAIR in NSCLC patients is related to cisplatin resistance (88). In A549 cells, lncRNA-HOTAIR increases cisplatin resistance by decreasing p21 expression and activating the wingless/integrated (Wnt) signaling pathway (89). In NSCLC patients, lncRNA-MEG3 enhances cisplatin sensitivity by regulating the miR-21-5p/SOX7 axis (90). The lncRNA-AK126698 regulates cisplatin resistance through the classic Wnt signaling pathway. Knockdown of the lncRNA-AK126698 will lead to activation of the Wnt/β-catenin pathway and the inhibition of apoptosis (91). The expression level of lncRNA-MEG3 is low in LAD tissues that are insensitive to cisplatin. In addition, chemotherapy based on cisplatin is less effective in patients with low lncRNA-MEG3 expression levels (92). It has been reported that the lncRNA-XIST modulates transforming growth factor-β (TGF-β) signaling by directly interacting with SMAD2, which impacts apoptosis, development of cisplatin (DDP)-mediated apoptosis, and resistance to DDP in NSCLC cells (93). The above research shows that lncRNAs may provide a novel treatment method and improve the prognosis of lung cancer patients; however, further research is needed for clinical application.
LncRNA and EGFR-TKI resistance
EGFR-TKIs are the first-line treatment for lung cancer patients with EGFR gene mutations. LncRNAs play a key role in the gefitinib resistance of lung cancer cells by regulating cell proliferation and apoptosis. The expression of the lncRNA-UCA1 in lung cancer is up-regulated and EGFR-TKI resistance is induced. lncRNA-UCA1 regulates EGFR-TKI resistance by activating the protein kinase B/mammalian target of rapamycin (AKT/mTOR) pathway and EMT, thereby affecting the prognosis of patients (94). The lncRNA-GAS5 increases the sensitivity of lung cancer cells to EGFR-TKIs by regulating the EGFR signaling pathway and IGF-1R (57). Overexpression of the lncRNA-MIR31HG can reduce the sensitivity of NSCLC cell lines to gefitinib and activate the EGFR/phosphatidylinositol 3 kinase (PI3K)/AKT pathway to block cell proliferation and cell cycle arrest in the G0/G1 phase (95). The expression of lncRNA-CASC9 is up-regulated in PC9-gefitinib resistant cells (PC9G), and knockdown of lncRNA-CASC9 will increase the sensitivity of PC9G cells to gefitinib (96). The expression level of EWAST1 (Linc00227) is low in PC9G cells, and overexpression of EWAST1 increases the sensitivity of PC9G cells to gefitinib (96). The lncRNA-BC087858 can induce EGFR-TKI resistance via EMT. The overexpression of lncRNA-BC087858 is associated with poor prognosis in NSCLC patients (97). Up-regulated lncRNA-HOTAIR can restore gefitinib sensitivity in gefitinib-resistant cells (PC9/R, H1299, and A549) by inducing cell apoptosis and activating EMT (98). Differentially-expressed lncRNAs are related to drug resistance in lung cancer cells, which can predict the drug treatment response of lung cancer patients and hopefully become a clinical biomarker for diagnosis and prognostic evaluation.
LncRNA and radio sensitivity
Radiotherapy is crucial for most patients with lung cancer, especially for those with advanced lung cancer (99). Studies have shown that lncRNAs are involved in radiation-induced DNA damage, suggesting that lncRNAs can regulate the sensitivity of cells to radiotherapy (100). Knockout of the lncRNA-PVT1 gene can enhance the radiosensitivity of NSCLC by inhibiting the expression of miR-195 (101). LncRNA-GAS5 can inhibit the expression of miR-135b, thereby inhibiting tumorigenesis and enhancing radiosensitivity (60). LINC00483/miR-144 regulates the radiosensitivity and EMT of LAD by interacting with HOXA10 (102). The above research shows that lncRNAs can help to predict the effectiveness of radiotherapy in patients. At present, the detailed mechanism of lncRNA resistance to radiotherapy requires further study.
Prospect of lncRNAs application for lung cancer in the future
LncRNAs application caused attractive concern as novel biomarkers for lung cancer diagnosis and prediction while there are also some challenges existed.
Firstly, the use of lncRNAs as the potential biomarkers for lung cancer diagnosis and prediction was in the stage of preclinic research for its infancy. Numerous functions of lncRNAs still need large validation and further explorations while scientific and clinic studies are required before the successful clinical translation is set. As one of the ncRNAs, a growing body of researches aim to investigate the biological roles of lncRNAs in tumorigenesis and the interaction with other types of ncRNAs such as miRNA, circRNA. These interacted effects may provide information on the assistant prognosis and prediction for lung cancer which also need further elucidation.
Secondly, lncRNA caused wide concern for its high stability and easy detection in body fluid. However, studies have reported an instability in special extreme conditions (103,104). Standard detected techniques and conditions were necessary to maintain the samples stability.
Thirdly, various lncRNAs and their functions were reported in recent studies. The combination of different lncRNAs formed a system of diagnosis and perdition in lung cancer which might produce a collaborated contribution other than the preserved specific effect of each individual. Expanded number and diversity of clinical samples were needed to improve the specificity and sensitivity in the future research.
Conclusions
We performed a comprehensive review of the published literature focusing on the significantly differentially-expressed lncRNAs in lung cancer tissues compared with healthy individuals and normal tissues. The expression of lncRNA is stable in body fluids and exhibits tissue specificity. As a non-invasive detection method, lncRNA detection can greatly reduce the pain of patients in diagnosis and postoperatively, as compared with traditional tissue biopsy. Therefore, differentially-expressed lncRNAs in external circulation are expected to become biomarkers for lung cancer diagnosis and a potential therapeutic target. In addition, differentially-expressed lncRNAs are involved in regulating drug resistance and radiosensitivity, which can be used to predict the sensitivity of patients to chemotherapy and targeted therapy. At present, the mechanism of lncRNA regulation of the occurrence and development of lung cancer requires further study, and selective targeting of lncRNAs is expected to become a novel therapeutic approach for lung cancer.
Acknowledgments
Funding: This article was funded by the Bethune Charitable Foundation Pharmaceutical Research Capacity Building Project (No. B-19-H-0100622).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-897/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-897/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Suster DI, Mino-Kenudson M. Molecular Pathology of Primary Non-small Cell Lung Cancer. Arch Med Res 2020;51:784-98. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol 2015;10:1240-2. [Crossref] [PubMed]
- Vijayvergia N, Mehra R. Clinical challenges in targeting anaplastic lymphoma kinase in advanced non-small cell lung cancer. Cancer Chemother Pharmacol 2014;74:437-46. [Crossref] [PubMed]
- Jiang L, Li Z, Wang R. Long non-coding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications Int J Oncol 2019;55:585-96. (Review). [Crossref] [PubMed]
- Sun R, Wang R, Chang S, et al. Long Non-Coding RNA in Drug Resistance of Non-Small Cell Lung Cancer: A Mini Review. Front Pharmacol 2019;10:1457. [Crossref] [PubMed]
- Inage T, Nakajima T, Yoshino I, et al. Early Lung Cancer Detection. Clin Chest Med 2018;39:45-55. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Wang C, Jiang Y, Lei Q, et al. Potential Diagnostic and Prognostic Biomarkers of Circular RNAs for Lung Cancer in China. Biomed Res Int 2019;2019:8023541. [Crossref] [PubMed]
- Wang H, Li M, Wang Y, et al. Construction of a Nomogram Based on lncRNA and Patient's Clinical Characteristics to Improve the Prognosis of Non-Small Cell Lung Cancer. Technol Cancer Res Treat 2022;21:15330338221097215. [Crossref] [PubMed]
- Peng W, Wang J, Shan B, et al. Diagnostic and Prognostic Potential of Circulating Long Non-Coding RNAs in Non Small Cell Lung Cancer. Cell Physiol Biochem 2018;49:816-27. [Crossref] [PubMed]
- Liu Y, Wang L, Liu H, et al. The Prognostic Significance of Metabolic Syndrome and a Related Six-lncRNA Signature in Esophageal Squamous Cell Carcinoma. Front Oncol 2020;10:61. [Crossref] [PubMed]
- Miao R, Ge C, Zhang X, et al. Combined eight-long noncoding RNA signature: a new risk score predicting prognosis in elderly non-small cell lung cancer patients. Aging (Albany NY) 2019;11:467-79. [Crossref] [PubMed]
- Xie W, Yuan S, Sun Z, et al. Long noncoding and circular RNAs in lung cancer: advances and perspectives. Epigenomics 2016;8:1275-87. [Crossref] [PubMed]
- Gong Y, Zhu W, Sun M, et al. Bioinformatics Analysis of Long Non-coding RNA and Related Diseases: An Overview. Front Genet 2021;12:813873. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Jiang L, Wang R, Fang L, et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics 2019;9:2460-74. [Crossref] [PubMed]
- Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv Exp Med Biol 2017;1008:47-74. [Crossref] [PubMed]
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol 2019;21:542-51. [Crossref] [PubMed]
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [Crossref] [PubMed]
- Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 2018;34:142-57. [Crossref] [PubMed]
- Lin C, Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol 2018;28:287-301. [Crossref] [PubMed]
- Ming H, Li B, Zhou L, et al. Long non-coding RNAs and cancer metastasis: Molecular basis and therapeutic implications. Biochim Biophys Acta Rev Cancer 2021;1875:188519. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Azizi Z, Mirtavoos-Mahyari H, Karimi R, et al. Long non-coding RNAs: Diverse roles in various disorders. Hum Antibodies 2019;27:221-5. [Crossref] [PubMed]
- Thai P, Statt S, Chen CH, et al. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol 2013;49:204-11. [Crossref] [PubMed]
- Li P, Zhang N, Ping F, et al. lncRNA SCAL1 inhibits inducible nitric oxide synthase in lung cells under high-glucose conditions. Exp Ther Med 2019;18:1831-6. [Crossref] [PubMed]
- Li S, Mei Z, Hu HB, et al. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol 2018;233:6679-88. [Crossref] [PubMed]
- Tang Y, Xiao G, Chen Y, et al. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer Drugs 2018;29:725-35. [Crossref] [PubMed]
- Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 2011;6:1984-92. [Crossref] [PubMed]
- Lin L, Li H, Zhu Y, et al. Expression of metastasis-associated lung adenocarcinoma transcript 1 long non-coding RNA in vitro and in patients with non-small cell lung cancer. Oncol Lett 2018;15:9443-9. [Crossref] [PubMed]
- Liu X, Huang G, Zhang J, et al. Prognostic and clinicopathological significance of long noncoding RNA MALAT-1 expression in patients with non-small cell lung cancer: A meta-analysis. PLoS One 2020;15:e0240321. [Crossref] [PubMed]
- Wang D, Zhang S, Zhao M, et al. LncRNA MALAT1 accelerates non-small cell lung cancer progression via regulating miR-185-5p/MDM4 axis. Cancer Med 2020;9:9138-49. [Crossref] [PubMed]
- Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther 2015;14:268-77. [Crossref] [PubMed]
- Naemura M, Murasaki C, Inoue Y, et al. Long Noncoding RNA ANRIL Regulates Proliferation of Non-small Cell Lung Cancer and Cervical Cancer Cells. Anticancer Res 2015;35:5377-82. [PubMed]
- Xie Y, Zhang Y, Du L, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol 2018;12:648-58. [Crossref] [PubMed]
- Lu Y, Zhou X, Xu L, et al. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther 2016;9:3077-84. [PubMed]
- Lin L, Gu ZT, Chen WH, et al. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol 2015;10:14. [Crossref] [PubMed]
- Hu X, Bao J, Wang Z, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol 2016;37:3497-504. [Crossref] [PubMed]
- Huang Z, Lei W, Hu HB, et al. H19 promotes non-small-cell lung cancer (NSCLC) development through STAT3 signaling via sponging miR-17. J Cell Physiol 2018;233:6768-76. [Crossref] [PubMed]
- Zhang Q, Li X, Li X, et al. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J Cell Biochem 2018;119:4447-57. [Crossref] [PubMed]
- Ren J, Fu J, Ma T, et al. LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle 2018;17:1372-80. [Crossref] [PubMed]
- Zhou Y, Sheng B, Xia Q, et al. Association of long non-coding RNA H19 and microRNA-21 expression with the biological features and prognosis of non-small cell lung cancer. Cancer Gene Ther 2017;24:317-24. [Crossref] [PubMed]
- Sun W, Zhang L, Yan R, et al. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. Onco Targets Ther 2019;12:3945-54. [Crossref] [PubMed]
- Huang Y, Ni R, Wang J, et al. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother 2019;109:1851-9. [Crossref] [PubMed]
- Guo D, Wang Y, Ren K, et al. Knockdown of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer by regulating miR-497 expression. Exp Cell Res 2018;362:172-9. [Crossref] [PubMed]
- Chen W, Zhu H, Yin L, et al. lncRNA-PVT1 Facilitates Invasion Through Upregulation of MMP9 in Nonsmall Cell Lung Cancer Cell. DNA Cell Biol 2017;36:787-93. [Crossref] [PubMed]
- Yang YR, Zang SZ, Zhong CL, et al. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol 2014;7:6929-35. [PubMed]
- Huang C, Liu S, Wang H, et al. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res 2016;8:5025-34. [PubMed]
- Pan Y, Liu L, Cheng Y, et al. Amplified LncRNA PVT1 promotes lung cancer proliferation and metastasis by facilitating VEGFC expression. Biochem Cell Biol 2020;98:676-82. [Crossref] [PubMed]
- Qiu C, Li S, Sun D, et al. lncRNA PVT1 accelerates progression of non-small cell lung cancer via targeting miRNA-526b/EZH2 regulatory loop. Oncol Lett 2020;19:1267-72. [PubMed]
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014;7:90. [Crossref] [PubMed]
- Chen L, Huang S, Huang J, et al. Role and Mechanism of Exosome-Derived Long Noncoding RNA HOTAIR in Lung Cancer. ACS Omega 2021;6:17217-27. [Crossref] [PubMed]
- Zhang C, Xu L, Deng G, et al. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci China Life Sci 2020;63:1265-8. [Crossref] [PubMed]
- Li H, Cui Z, Lv X, et al. Long Non-coding RNA HOTAIR Function as a Competing Endogenous RNA for miR-149-5p to Promote the Cell Growth, Migration, and Invasion in Non-small Cell Lung Cancer. Front Oncol 2020;10:528520. [Crossref] [PubMed]
- Agulló-Ortuño MT, Díaz-García CV, Agudo-López A, et al. Relevance of insulin-like growth factor 1 receptor gene expression as a prognostic factor in non-small-cell lung cancer. J Cancer Res Clin Oncol 2015;141:43-53. [Crossref] [PubMed]
- Dong S, Qu X, Li W, et al. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol 2015;8:43. [Crossref] [PubMed]
- Zhang N, Yang GQ, Shao XM, et al. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci 2016;20:2271-7. [PubMed]
- Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol 2019;234:20721-7. [Crossref] [PubMed]
- Xue Y, Ni T, Jiang Y, et al. Long Noncoding RNA GAS5 Inhibits Tumorigenesis and Enhances Radiosensitivity by Suppressing miR-135b Expression in Non-Small Cell Lung Cancer. Oncol Res 2017;25:1305-16. [Crossref] [PubMed]
- Ma J, Miao H, Zhang H, et al. LncRNA GAS5 modulates the progression of non-small cell lung cancer through repressing miR-221-3p and up-regulating IRF2. Diagn Pathol 2021;16:46. [Crossref] [PubMed]
- Zhu L, Zhou D, Guo T, et al. LncRNA GAS5 inhibits Invasion and Migration of Lung Cancer through influencing EMT process. J Cancer 2021;12:3291-8. [Crossref] [PubMed]
- Nishikawa S, Menju T, Takahashi K, et al. Statins may have double-edged effects in patients with lung adenocarcinoma after lung resection. Cancer Manag Res 2019;11:3419-32. [Crossref] [PubMed]
- Al-Rugeebah A, Alanazi M, Parine NR. MEG3: an Oncogenic Long Non-coding RNA in Different Cancers. Pathol Oncol Res 2019;25:859-74. [Crossref] [PubMed]
- Lu KH, Li W, Liu XH, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013;13:461. [Crossref] [PubMed]
- Wu JL, Meng FM, Li HJ. High expression of lncRNA MEG3 participates in non-small cell lung cancer by regulating microRNA-7-5p. Eur Rev Med Pharmacol Sci 2018;22:5938-45. [PubMed]
- Yang Z, Wang Z, Duan Y. LncRNA MEG3 inhibits non-small cell lung cancer via interaction with DKC1 protein. Oncol Lett 2020;20:2183-90. [Crossref] [PubMed]
- Wang C, Tao X, Wei J. Effects of LncRNA MEG3 on immunity and autophagy of non-small cell lung carcinoma through IDO signaling pathway. World J Surg Oncol 2021;19:244. [Crossref] [PubMed]
- Jiang W, Zhang D, Xu B, et al. Long non-coding RNA BANCR promotes proliferation and migration of lung carcinoma via MAPK pathways. Biomed Pharmacother 2015;69:90-5. [Crossref] [PubMed]
- Yang L, Liu G. lncRNA BANCR suppresses cell viability and invasion and promotes apoptosis in non-small-cell lung cancer cells in vitro and in vivo. Cancer Manag Res 2019;11:3565-74. [Crossref] [PubMed]
- Urbanska EM, Sørensen JB, Melchior LC, et al. Changing ALK-TKI-Resistance Mechanisms in Rebiopsies of ALK-Rearranged NSCLC: ALK- and BRAF-Mutations Followed by Epithelial-Mesenchymal Transition. Int J Mol Sci 2020;21:2847. [Crossref] [PubMed]
- Sun M, Liu XH, Wang KM, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 2014;13:68. [Crossref] [PubMed]
- Zhang DY, Zou XJ, Cao CH, et al. Identification and Functional Characterization of Long Non-coding RNA MIR22HG as a Tumor Suppressor for Hepatocellular Carcinoma. Theranostics 2018;8:3751-65. [Crossref] [PubMed]
- Su W, Feng S, Chen X, et al. Silencing of Long Noncoding RNA MIR22HG Triggers Cell Survival/Death Signaling via Oncogenes YBX1, MET, and p21 in Lung Cancer. Cancer Res 2018;78:3207-19. [Crossref] [PubMed]
- Li DS, Ainiwaer JL, Sheyhiding I, et al. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci 2016;20:2285-95. [PubMed]
- Zhang C, Su C, Song Q, et al. LncRNA PICART1 suppressed non-small cell lung cancer cells proliferation and invasion by targeting AKT1 signaling pathway. Am J Transl Res 2018;10:4193-201. [PubMed]
- Zhao JM, Cheng W, He XG, et al. Long non-coding RNA PICART1 suppresses proliferation and promotes apoptosis in lung cancer cells by inhibiting JAK2/STAT3 signaling. Neoplasma 2018;65:779-89. [Crossref] [PubMed]
- Zhang L, Wang Y, Xia S, et al. Long noncoding RNA PANDAR inhibits the development of lung cancer by regulating autophagy and apoptosis pathways. J Cancer 2020;11:4783-90. [Crossref] [PubMed]
- Han L, Zhang EB, Yin DD, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis 2015;6:e1665. [Crossref] [PubMed]
- Tang W, Li X, Xie X, et al. EGFR inhibitors as adjuvant therapy for resected non-small cell lung cancer harboring EGFR mutations. Lung Cancer 2019;136:6-14. [Crossref] [PubMed]
- Zhou J, Hu Q, Zhang X, et al. Sensitivity to chemotherapeutics of NSCLC cells with acquired resistance to EGFR-TKIs is mediated by T790M mutation or epithelial-mesenchymal transition. Oncol Rep 2018;39:1783-92. [Crossref] [PubMed]
- Bermúdez M, Aguilar-Medina M, Lizárraga-Verdugo E, et al. LncRNAs as Regulators of Autophagy and Drug Resistance in Colorectal Cancer. Front Oncol 2019;9:1008. [Crossref] [PubMed]
- Song Y, Zou L, Li J, et al. LncRNA SNHG8 promotes the development and chemo-resistance of pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci 2018;22:8161-8. [PubMed]
- Gao J, Meng Q, Zhao Y, et al. EHD1 confers resistance to cisplatin in non-small cell lung cancer by regulating intracellular cisplatin concentrations. BMC Cancer 2016;16:470. [Crossref] [PubMed]
- Liao S, Yu C, Liu H, et al. Long non-coding RNA H19 promotes the proliferation and invasion of lung cancer cells and regulates the expression of E-cadherin, N-cadherin, and vimentin. Onco Targets Ther 2019;12:4099-107. [Crossref] [PubMed]
- Wang Q, Cheng N, Li X, et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget 2017;8:2558-67. [Crossref] [PubMed]
- Fang Z, Chen W, Yuan Z, et al. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed Pharmacother 2018;101:536-42. [Crossref] [PubMed]
- Liu MY, Li XQ, Gao TH, et al. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J Thorac Dis 2016;8:3314-22. [Crossref] [PubMed]
- Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One 2013;8:e77293. [Crossref] [PubMed]
- Wang P, Chen D, Ma H, et al. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. Onco Targets Ther 2017;10:5137-49. [Crossref] [PubMed]
- Yang Y, Li H, Hou S, et al. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One 2013;8:e65309. [Crossref] [PubMed]
- Liu J, Wan L, Lu K, et al. The Long Noncoding RNA MEG3 Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma. PLoS One 2015;10:e0114586. [Crossref] [PubMed]
- Xu X, Zhou X, Chen Z, et al. Silencing of lncRNA XIST inhibits non-small cell lung cancer growth and promotes chemosensitivity to cisplatin. Aging (Albany NY) 2020;12:4711-26. [Crossref] [PubMed]
- Cheng N, Cai W, Ren S, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015;6:23582-93. [Crossref] [PubMed]
- Wang B, Jiang H, Wang L, et al. Increased MIR31HG lncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol Lett 2017;13:3494-500. [Crossref] [PubMed]
- Ma P, Zhang M, Nie F, et al. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother 2017;87:20-6. [Crossref] [PubMed]
- Pan H, Jiang T, Cheng N, et al. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget 2016;7:49948-60. [Crossref] [PubMed]
- Wang Q, Li X, Ren S, et al. HOTAIR induces EGFR-TKIs resistance in non-small cell lung cancer through epithelial-mesenchymal transition. Lung Cancer 2020;147:99-105. [Crossref] [PubMed]
- Wu AJ, Garay E, Foster A, et al. Definitive Radiotherapy for Local Recurrence of NSCLC After Surgery. Clin Lung Cancer. 2017;18:e161-e8. [Crossref] [PubMed]
- Li L, Zhu T, Gao YF, et al. Targeting DNA Damage Response in the Radio(Chemo)therapy of Non-Small Cell Lung Cancer. Int J Mol Sci 2016;17:839. [Crossref] [PubMed]
- Wu D, Li Y, Zhang H, et al. Knockdown of Lncrna PVT1 Enhances Radiosensitivity in Non-Small Cell Lung Cancer by Sponging Mir-195. Cell Physiol Biochem 2017;42:2453-66. [Crossref] [PubMed]
- Yang QS, Li B, Xu G, et al. Long noncoding RNA LINC00483/microRNA-144 regulates radiosensitivity and epithelial-mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J Cell Physiol 2019;234:11805-21. [Crossref] [PubMed]
- Arita T, Ichikawa D, Konishi H, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res 2013;33:3185-93. [PubMed]
- Chen Y, Zitello E, Guo R, et al. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin Transl Med 2021;11:e367. [Crossref] [PubMed]


