
Screening of atrial fibrillation diagnostic markers based on a GEO database chip and bioinformatics analysis
Highlight box
Key findings
• We identified 5 potential diagnostic key genes GPR22, COG5, GALNT16, OTOGL, and MCOLN3 for AF.
What is known and what is new?
• To date, many clues suggests a close association between inflammation and the development of AF.
• But the significant biomarkers between inflammation and AF do not have been well classified, present study identified GPR22, COG5, GALNT16, OTOGL, and MCOLN3 as the critical inflammation regulators for AF.
What is the implication, and what should change now?
• GPR22, COG5, GALNT16, OTOGL, and MCOLN3 may represent potential therapeutic targets for AF. A resulting risk model was constructed to show the predictive ability with these genes for diagnosing AF.
Introduction
Atrial fibrillation (AF) is one of the most common clinical arrhythmias, AF can easily lead to complications, such as thromboembolism, heart failure and stroke, and can seriously affect patients’ quality of life and safety. In a recent study, an analysis of electrocardiograms of middle-aged adults showed that 250 of every 100,000 people suffer from AF, China ranks 3rd in terms of the incidence of AF, and due to the aging population, the incidence of AF continues to increase (1-3).
Study on the etiology of familial AF have shown that AF has a certain heredity (4). Similarly, etiological studies of patients with isolated AF without organic or systemic disease confirmed that AF is a disease of a genetic nature (5,6). A large meta-analysis of >60,000 AF patients and >500,000 reference subjects identified 97 genetic loci that were significantly associated with AF, and a genetic analysis showed that genetic variants among them accounted for 42% of the heritability of AF (7). All of these studies suggest that genetic factors may play an important role in the development of AF.
Epidemiological findings suggest that smoking, obesity, hypertension, and obstructive sleep apnea are risk factors for AF (8-10). Recent research suggests that inflammatory markers, such as interleukin 6, tumor necrosis factor-α, and myeloperoxidase), are positively associated with AF progression and predict AF outcomes (11-13). Several clinical studies have shown that inflammatory factors can predict the onset and prognosis of AF and may be able to be used as biomarkers of AF (14,15). The above evidence suggests a close association between inflammation and the development of AF. However, there are few reports on the potential pathogenesis and biomarkers of AF.
In recent years, following the application of molecular biology and cellular electrophysiology techniques, great advances have been made in determining the molecular genetic etiology of AF, and while more and more pathogenic genes have been identified, the relationship between these genes and AF is still not completely clear. Thus, this study sought to identify the causative genes of AF and their relationship with the pathogenesis of AF, and to explore the relationship and role of inflammation-associated immune cells in AF. Our findings may help in the early detection of individuals at high risk of AF, provide a theoretical basis for finding new targets for AF, facilitate the early diagnosis and treatment of AF, improve clinical outcomes, and reduce the burden and suffering of patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1457/rc).
Methods
Data collection and normalization
We downloaded the AF and control data sets from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE41177 and GSE115574 data sets were used in the follow-up study. The data sets were based on the GPL570 microarray platform, and R software (version 3.6.3) was employed to normalize the data using the “limma” package. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of DEGs
We used the limma package of R software to screen the normalized data and identify the differentially expressed genes (DEGs). DEGs with a fold-change value >0.5 and an adjusted P value <0.5 were considered statistically significant DEGs. Volcano maps were drawn for the DEGs using the ggplot2 package. The volcano maps were visualized by pheatmap maps for the DEGs and visualized using the venny tool Wayne plots (version 2.1, https://bioinfogp.cnb.csic.es/tools/venny/). The core DEGs were identified, undefined genes were removed, and the remaining core DEGs were included in the next step of the study.
Protein-protein interaction (PPI) and gene-gene interaction (GGI) network analyses
A GGI network of the DEGs was constructed using data from the Genemania (http://genemania.org/) database. A PPI network was constructed using the STRING (https://string-db.org) database, and a medium confidence was (0.400).
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
The core genes were analyzed using the clusterProfiler package in R language for the GO and KEGG analysis. A P value <0.05 indicated significant functional enrichment. The GO analysis revealed the potential biological functions in terms of the biological processes, cellular compositions, and molecular functions, and the KEGG analysis was used to analyze the potential mechanisms of the core gene action.
Gene set enrichment analysis (GSEA) analysis
The core genes and their log fold-change values were analyzed by a GSEA through clusterProfiler and ReactomePA in R language, and the top 5 GO and KEGG results were visualized by C2 and C5 according to the adjusted P values from low to high.
Diagnostic model
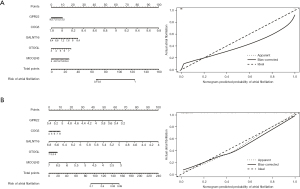
The pROC package in R language was used to construct the receiver operating characteristic (ROC) curves to determine the area under the curves (AUCs) of the core genes to assess the diagnostic efficacy of the core genes for the diagnosis of AF. Based on the AUCs, the top 5 core genes in the 2 data sets were selected as diagnostic factors and a nomogram was constructed. We used GSE115574 dataset as the training set and GSE41177 as the validation set to evaluate the stability of the model. The accuracy of the model was assessed based on the concordance index (C-index), and the correction curve was plotted to evaluate the performance of the model.
Immune infiltration
After normalization, the data from the GSE41177 data set were imported into the CIBERSORT website (https://cibersort.stanford.edu/). The expression matrix of human immune cell subtypes was deconvolved, and the proportion of 22 kinds of immune cells was obtained by 500 permutation calculations. The results were visualized using R language.
Immunological correlation analysis
The core genes in the GSE41177 data set and the genes calculated by CIBERSORT were subjected to a correlation analysis, and the results were visualized using the R language psych package to assess the potential regulatory roles of the core genes on the immune cells.
Regulatory gene miRNA prediction
The regulatory micro ribonucleic acids (miRNAs) for the diagnostic factors were obtained from the FunRich (http://www.funrich.org/, version 3.1.3). The data were visualized using Cytoscape.
Statistical analysis
The pheatmap package was applied to construct the expression heat map of important genes in AF and control. Statistical tests were performed using the R language limma package to compare the differences in expression of important genes in AF and control, and P<0.05 was considered a statistically significant difference.
Results
Data pre-processing and DEG identification
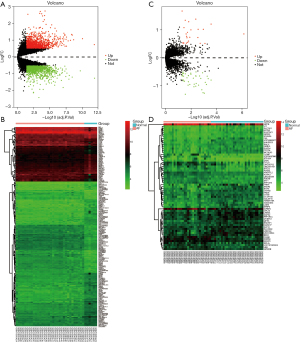
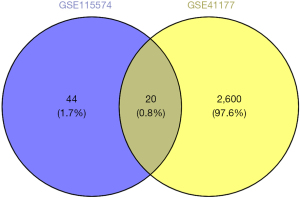
The present study included 2 data sets (i.e., GSE41177 and GSE115574) that were normalized separately (Figure S1). The results of the volcano plot and cluster analyses indicated that the DEGs were more significant in the GSE41177 data set (Figure 1). Subsequently, we analyzed the gene expression matrixes of these 2 data sets and identified a total of 20 DEGs (Figure 2).
Analysis of PPIs and GGIs
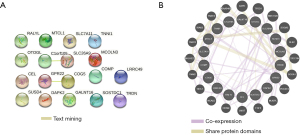
The PPI network analysis of the 20 DEGs by the STRING database revealed that GPR22 was closely related to COG5 (Figure 3A). The GGI network analysis revealed that the DEGs interacted with 19 genes (Figure 3B).
DEG enrichment analysis
We conducted a GO analysis to examine the main functions of the DEGs. The biological processes mainly involved muscle contraction, post-synaptic membrane organization, and sulfur compound transport. The cellular molecular functions mainly involved bone morphogenetic protein (BMP) binding, anon anti-porter activity, and sulfur compound transmigration. The molecular functions mainly involved BMP binding, anon anti-porter activity and sulfur compound transmembrane transporter activity (Figure 4A).
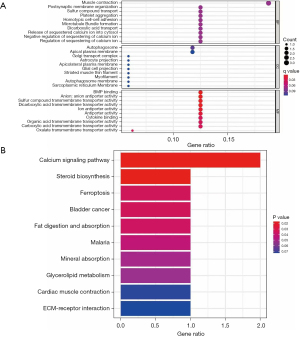
The subsequent KEGG analysis showed that the DEGs focused on the calcium signaling pathway, ferroptosis, and the extracellular matrix–receptor interaction (Figure 4B). Due to the differences in the DEGs between the 2 data sets, we analyzed the functions of the DEGs in the data sets separately. In the GSE41177 data set, the DEGs were mainly involved in molecular function, binding, and metabolic process (Figure 5A). Conversely, in the GSE115574 data set, the DEGs were mainly involved in cellular components, the metabolic process, and biological regulation (Figure 5B).

Identification of key genes and risk model construction
We evaluated the diagnostic efficacy of the DEGs in both data sets (Figures S2-S4), and identified a total of 5 core genes; that is, GPR22, COG5, GALNT16, OTOGL, and MCOLN3 (Figure 6). In the training set, the correction C-index of the constructed model was 0.99, while in the validation set, the correction C-index of the model was also 0.94, indicating that our model has high stability and accuracy.
Immune infiltration analysis
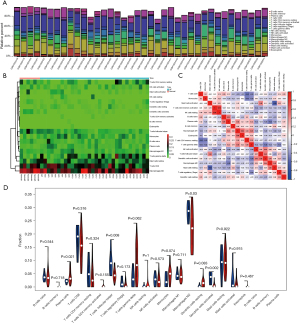
We analyzed the abundance of immune infiltration in the GSE41177 data, and the results showed that M1 macrophages, gamma delta T cells, resting mast cells, cluster of differentiation (CD) 8 T cells, and M2 macrophages were more variable in the AF patients than the normal patients (Figure 7). Additionally, plasma cells and M2 macrophages were significantly more increased in the AF patients than the normal patients, and follicular helper T cells and activated dendritic cells were significantly more decreased in the AF patients than the normal patients (Figure 7).
Functional analysis of model genes
We predicted the targeting miRNAs of the model genes, and found that GPR22 and OTOGL had the most potential miRNAs, especially GPR22, which had 14 miRNAs, while COG5 and MCOLN3 had no potential miRNAs (Figure 8A). We then analyzed the correlations between these model genes and immune cells and found that GPR22 was positively correlated with M2 macrophages and CD4 memory resting T cells, COG5 was positively correlated with M2 macrophages, activated dendritic cells, and mast cells, and GALNT16 was negatively correlated with plasma cells (Figure 8B).

Discussion
Research has shown that the ectopic origin site of AF is mainly located in the left atrium, while the right atrium is only involved in the formation of foldback, and the degree of structural remodeling of the left auricle and the flow rate are significantly correlated with post-operative AF recurrence (16-19). Thus, the systematic study of left heart tissue in AF patients is particularly important.
In our study, an analysis of the high-throughput sequencing results of the left atrium revealed 20 DEGs in AF and normal atrial tissue, and an enrichment analysis of the functions of these genes revealed that certain DEGs are involved in regulating muscle contraction and the calcium signaling pathway, and are closely associated with immune cells. Subsequently, we constructed a prediction model comprising 5 genes that could predict the occurrence of AF.
Electrical remodeling is the most common alteration in AF, and occurs as a result of a decrease in L-type calcium current conductance and an increase in inward rectifier current conductance (20). Molina et al. suggested that AF is associated with calcium overload and that abnormal calcium plays an important role in increasing patients’ susceptibility to AF in various models of heart failure (21). A large body of evidence from recent studies suggests that intracellular calcium ions play an important role in the development and maintenance of AF (22,23). It has been inferred that the calcium signaling pathway plays an important role in the development of AF, but the specific regulatory mechanisms are not yet clear.
Ferroptosis, a novel form of cell death regulation caused by the accumulation of iron-triggered lipid peroxidation, has recently been revealed to play a key role in the pathogenesis of various cardiovascular diseases and the existence of potential therapeutic targets (24,25). Conversely, studies of AF have shown that the inhibition of iron death reduces patients’ susceptibility to frequent excessive alcohol consumption-induced AF (26). Additionally, iron transporter protein-mediated iron death has been shown to be associated with the new-onset AF in lipopolysaccharide-induced endotoxemia (27). In our study, the KEGG results suggested that the identified DEGs may be associated with the above pathways, and the current study yielded similar findings, providing further evidence of the key role of these DEGs in AF.
A number of risk models constructed from GPR22, COG5, GALNT16, OTOGL, but MCOLN3, OTOGL has not been reported and thus should be the focus of subsequent studies. GPR22 is a member of the G protein-coupled receptor 1 family and is selectively highly expressed in the brain and heart (28-30). Research has shown that expression level of GPR22 may be associated with heart failure progression and thus is a potential therapeutic target of AF (28-30). Conversely, COG5 and GALNT16 have been shown to be associated with atheromatous plaque and lipid metabolism, respectively, and are involved in the regulation of cardiovascular disease (31,32). The mucin subfamily is also known as transient receptor potential channels, is highly expressed in cardiac fibroblasts, and MCOLN3, a member of the mucin subfamily, plays an important role in the regulation of calcium ion homeostasis (33). The downregulation of MCOLN3 has been shown to be closely associated with AF related to chronic primary mitral valve closure insufficiency (34).
Recent studies have revealed that immune cell populations are involved in the development of AF and that cytokines released by lymphocytes can affect the conduction system of the heart, especially with a greater potential effect on AF (35). T lymphocytes and their subpopulations are important factors in the body’s immunity system and are involved in regulating the inflammatory response, thus affecting the maintenance and progression of AF (36).
A study has shown that the number of CD45+ and CD3+ cells is significantly elevated in the atrial adipose tissue of all AF patients, and that the degree of atrial inflammation affects the clinical prognosis of patients and may lead to a shift in the type of AF (37). Smorodinova et al. analyzed atrial myocardial tissue from 46 AF patients and showed that in addition to a significant increase in the number of both CD45+ and CD3+ cells, the number of inflammatory cells, such as macrophages and dendritic cells, was also significantly elevated (38).
In addition, the programmed cell death 1 (PD-1) and programmed cell death-Ligand 1 (PD-L1) pathways may play immunomodulatory roles in the pathogenesis of AF by regulating T cell excitation and thus promoting cytokine excretion (39). The above-mentioned findings suggest that the helper T cell population and its cytokines are closely associated with the progression of AF. Similar results were observed in our study, and GPR22, COG5, and GALNT16 were found to be closely associated with the expression of T lymphocyte subsets and macrophages.
In our present study, 5 potential key genes were identified by mining DEGs. These genes may represent potential therapeutic targets for AF. A resulting risk model was constructed to show the predictive ability. It should be noted that this study had some limitations. First, AF onset results from multiple genetic and environmental factors and co-actions that were not included in the analysis. Second, due to the lack of clinical samples, we were unable to perform gene expression validation, and more basic experiments need to be conducted to validate the functions and regulatory mechanisms of these genes. Third, the diagnosis model was based on a comparison between sinus rhythm and AF, which may have limitations in clinical application.
Conclusions
Our study identified a total of 5 potential key genes; that is, GPR22, COG5, GALNT16, OTOGL, and MCOLN3. Our findings may provide a theoretical basis for susceptibility analyses and target drug development in AF.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1457/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1457/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Andrade J, Khairy P, Dobrev D, et al. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res 2014;114:1453-68. [Crossref] [PubMed]
- Joseph PG, Healey JS, Raina P, et al. Global variations in the prevalence, treatment, and impact of atrial fibrillation in a multi-national cohort of 153 152 middle-aged individuals. Cardiovasc Res 2021;117:1523-31. [Crossref] [PubMed]
- Lippi G, Sanchis-Gomar F, Cervellin G. Global epidemiology of atrial fibrillation: An increasing epidemic and public health challenge. Int J Stroke 2021;16:217-21. [Crossref] [PubMed]
- Lubitz SA, Yin X, Fontes JD, et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA 2010;304:2263-9. [Crossref] [PubMed]
- Ragab AAY, Sitorus GDS, Brundel BBJJM, et al. The Genetic Puzzle of Familial Atrial Fibrillation. Front Cardiovasc Med 2020;7:14. [Crossref] [PubMed]
- Manoharan A, Sambandam R, Ballambattu VB. Genetics of atrial fibrillation-an update of recent findings. Mol Biol Rep 2022;49:8121-9. [Crossref] [PubMed]
- Roselli C, Chaffin MD, Weng LC, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225-33. [Crossref] [PubMed]
- Heijman J, Algalarrondo V, Voigt N, et al. The value of basic research insights into atrial fibrillation mechanisms as a guide to therapeutic innovation: a critical analysis. Cardiovasc Res 2016;109:467-79. [Crossref] [PubMed]
- Lau DH, Schotten U, Mahajan R, et al. Novel mechanisms in the pathogenesis of atrial fibrillation: practical applications. Eur Heart J 2016;37:1573-81. [Crossref] [PubMed]
- Turagam MK, Velagapudi P, Kocheril AG, et al. Commonly consumed beverages in daily life: do they cause atrial fibrillation? Clin Cardiol 2015;38:317-22. [Crossref] [PubMed]
- Li J, Solus J, Chen Q, et al. Role of inflammation and oxidative stress in atrial fibrillation. Heart Rhythm 2010;7:438-44. [Crossref] [PubMed]
- Rudolph V, Andrié RP, Rudolph TK, et al. Myeloperoxidase acts as a profibrotic mediator of atrial fibrillation. Nat Med 2010;16:470-4. [Crossref] [PubMed]
- Wu N, Xu B, Xiang Y, et al. Association of inflammatory factors with occurrence and recurrence of atrial fibrillation: a meta-analysis. Int J Cardiol 2013;169:62-72. [Crossref] [PubMed]
- Bruins P, te Velthuis H, Yazdanbakhsh AP, et al. Activation of the complement system during and after cardiopulmonary bypass surgery: postsurgery activation involves C-reactive protein and is associated with postoperative arrhythmia. Circulation 1997;96:3542-8. [Crossref] [PubMed]
- Zhang H, Li J, Chen X, et al. Association of Systemic Inflammation Score With Atrial Fibrillation: A Case-Control Study With Propensity Score Matching. Heart Lung Circ 2018;27:489-96. [Crossref] [PubMed]
- Di Biase L, Santangeli P, Natale A. How to ablate long-standing persistent atrial fibrillation? Curr Opin Cardiol 2013;28:26-35. [Crossref] [PubMed]
- Kanda T, Masuda M, Sunaga A, et al. Low left atrial appendage flow velocity predicts recurrence of atrial fibrillation after catheter ablation of persistent atrial fibrillation. J Cardiol 2015;66:377-81. [Crossref] [PubMed]
- Suksaranjit P, Marrouche NF, Han FT, et al. Relation of Left Atrial Appendage Remodeling by Magnetic Resonance Imaging and Outcome of Ablation for Atrial Fibrillation. Am J Cardiol 2018;122:83-8. [Crossref] [PubMed]
- Gonzalez-Casal D, Datino T, Soto N, et al. Anatomy of the left atrial appendage for the interventional cardiologist. Herzschrittmacherther Elektrophysiol 2022;33:195-202. [Crossref] [PubMed]
- Komal S, Yin JJ, Wang SH, et al. MicroRNAs: Emerging biomarkers for atrial fibrillation. J Cardiol 2019;74:475-82. [Crossref] [PubMed]
- Molina CE, Abu-Taha IH, Wang Q, et al. Profibrotic, Electrical, and Calcium-Handling Remodeling of the Atria in Heart Failure Patients With and Without Atrial Fibrillation. Front Physiol 2018;9:1383. [Crossref] [PubMed]
- Heijman J, Voigt N, Wehrens XH, et al. Calcium dysregulation in atrial fibrillation: the role of CaMKII. Front Pharmacol 2014;5:30. [Crossref] [PubMed]
- Wang X, Chen X, Dobrev D, et al. The crosstalk between cardiomyocyte calcium and inflammasome signaling pathways in atrial fibrillation. Pflugers Arch 2021;473:389-405. [Crossref] [PubMed]
- Hu H, Chen Y, Jing L, et al. The Link Between Ferroptosis and Cardiovascular Diseases: A Novel Target for Treatment. Front Cardiovasc Med 2021;8:710963. [Crossref] [PubMed]
- Huang F, Yang R, Xiao Z, et al. Targeting Ferroptosis to Treat Cardiovascular Diseases: A New Continent to Be Explored. Front Cell Dev Biol 2021;9:737971. [Crossref] [PubMed]
- Dai C, Kong B, Qin T, et al. Inhibition of ferroptosis reduces susceptibility to frequent excessive alcohol consumption-induced atrial fibrillation. Toxicology 2022;465:153055. [Crossref] [PubMed]
- Fang J, Kong B, Shuai W, et al. Ferroportin-mediated ferroptosis involved in new-onset atrial fibrillation with LPS-induced endotoxemia. Eur J Pharmacol 2021;913:174622. [Crossref] [PubMed]
- O'Dowd BF, Nguyen T, Jung BP, et al. Cloning and chromosomal mapping of four putative novel human G-protein-coupled receptor genes. Gene 1997;187:75-81. [Crossref] [PubMed]
- Lee J, Hever A, Willhite D, et al. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J 2005;19:1356-8. [Crossref] [PubMed]
- Adams JW, Wang J, Davis JR, et al. Myocardial expression, signaling, and function of GPR22: a protective role for an orphan G protein-coupled receptor. Am J Physiol Heart Circ Physiol 2008;295:H509-21. [Crossref] [PubMed]
- van der Laan SW, Siemelink MA, Haitjema S, et al. Genetic Susceptibility Loci for Cardiovascular Disease and Their Impact on Atherosclerotic Plaques. Circ Genom Precis Med 2018;11:e002115. [Crossref] [PubMed]
- Tabassum R, Rämö JT, Ripatti P, et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat Commun 2019;10:4329. [Crossref] [PubMed]
- Lelouvier B, Puertollano R. Mucolipin-3 regulates luminal calcium, acidification, and membrane fusion in the endosomal pathway. J Biol Chem 2011;286:9826-32. [Crossref] [PubMed]
- Çubukçuoğlu Deniz G, Durdu S, Doğan Y, et al. Molecular Signatures of Human Chronic Atrial Fibrillation in Primary Mitral Regurgitation. Cardiovasc Ther 2021;2021:5516185. [Crossref] [PubMed]
- Li T, Sun ZL, Xie QY. Meta-analysis Identifies Serum C-Reactive Protein as an Indicator of Atrial Fibrillation Risk After Coronary Artery Bypass Graft. Am J Ther 2016;23:e1586-96. [Crossref] [PubMed]
- Saigusa R, Winkels H, Ley K. T cell subsets and functions in atherosclerosis. Nat Rev Cardiol 2020;17:387-401. [Crossref] [PubMed]
- Wu L, Emmens RW, van Wezenbeek J, et al. Atrial inflammation in different atrial fibrillation subtypes and its relation with clinical risk factors. Clin Res Cardiol 2020;109:1271-81. [Crossref] [PubMed]
- Smorodinova N, Bláha M, Melenovský V, et al. Analysis of immune cell populations in atrial myocardium of patients with atrial fibrillation or sinus rhythm. PLoS One 2017;12:e0172691. [Crossref] [PubMed]
- Chang G, Chen Y, Liu Z, et al. The PD-1 with PD-L1 Axis Is Pertinent with the Immune Modulation of Atrial Fibrillation by Regulating T Cell Excitation and Promoting the Secretion of Inflammatory Factors. J Immunol Res 2022;2022:3647817. [Crossref] [PubMed]


