
Development and validation of a combined metabolism and immune prognostic model in lung adenocarcinoma
Highlight box
Key findings
• We constructed a stable comprehensive index of glycolysis and immunity (CIGI) prediction model.
What is known and what is new?
• The immune microenvironment and tumor metabolism, characterized by glycolysis, play a unique role in the progression of lung adenocarcinoma (LUAD).
• In this study, we established a new prognostic prediction model for LUAD patients that combines glycolysis ability and immune status. The CIGI score can be included as an independent prognostic factor for LUAD patients, and can also indicate the glycolytic ability and immune status of patients.
What is the implication, and what should change now?
• Our findings show that the CIGI model is very accurate as a prognostic tool for LUAD patients. In the future, we need to use prospective samples for research and conduct more in vivo and in vitro experiments for further research. And this model should be improved and applied to clinical treatment.
Introduction
Lung adenocarcinoma (LUAD) is one of the deadliest tumors and is also the main pathological subtype of lung cancer. Currently, the treatment of LUAD includes surgery, chemotherapy, targeted therapy, and other methods. The combined use of these treatments has led to great strides in terms of overall patient benefits; however, the 5-year survival rate for LUAD patients remains low. Thus, a new personalized treatment approach is urgently needed to improve the survival time of patients.
In recent years, immunotherapy, especially immune checkpoint blockade therapy, has been considered a major breakthrough in the treatment of LUAD and is widely regarded as one of the most promising cancer treatments (1). However, the efficacy of immune checkpoint inhibitors (ICIs) faces a problem common to other drug treatments; that of, individual differences of the patients. Some patients treated with ICIs have been found have longer overall survival (OS) than those who receive other treatments; however, there are still some patients who do not benefit from ICIs (2). Thus, further research on immune escape mechanisms needs to be conducted and potential therapeutic targets need to be identified to improve the therapeutic effect of immunotherapy and to identify beneficiary groups so that more patients can obtain better treatment outcomes.
The interplay between the metabolic changes of immune cells and tumor cells is considered a major critical point in tumor immune response and immunosuppression. Previous study has provided evidence that tumor metabolism not only plays a key role in tumor initiation and development by directly participating in energy metabolism but also affects the production and function of immune molecules by releasing metabolites, such as lactate (3). What’s more, the study has suggested that immune cells are regulated by metabolic reprogramming during differentiation, and the immune cells produced some biological effects, and this regulation is critical for immune responses (3). Thus, a comprehensive understanding of how the metabolic reprogramming of tumor cells modulates anti-tumor immune responses may lead to the development of therapeutic approaches that target metabolic pathways that may affect the efficacy of immunotherapy.
Tumor metabolic reprogramming involves multiple processes, of which glycolysis is the most prominent and typical feature, involves multiple processes, such as enhanced aerobic glycolysis, the increased utilization, uptake and consumption of glucose, and increased lipid and protein synthesis (4-6). As first observed by Warburg, even in the presence of large amounts of oxygen, tumor cells produce lactate in the cytoplasm primarily by metabolizing glucose, which is known as the “Warburg effect” or “aerobic glycolysis” (7). Recent studies have shown that the primary source of energy required for the activation of some immune cells, such as T cells, is glycolysis (8,9). At the same time, the products of glycolysis affect the action of immune cells. For example, lactate, one of the major metabolites of tumor cells, is often highly enriched in the tumor microenvironment, and can inhibit the cytolytic ability and immune surveillance ability of some T cells (10,11). It has also been observed that regulatory T cells (Tregs) may be resistant to lactate, and the lactate metabolic pathway may be activated in Tregs and is used to support cell metabolism, proliferation, and inhibitory function (10,11). This suggests that glycolysis plays a critical and complex role in immune cells and tumor cells. Despite the clear association between glycolysis and immune responses, few studies have closely examined this relationship.
In recent years, some studies have attempted to establish a prognostic model of lung adenocarcinoma. However, the current models are usually based on the whole genome and rarely focus on specific mechanisms. This makes it difficult for existing models to reflect the metabolic status and immune status of patients at the same time. The huge impact of tumor metabolism and tumor immunity on the prognosis and treatment of patients makes models based on glycolysis and immunity more targeted than other models. At the same time, this model also provides more information for follow-up treatment strategies. Therefore, a simple model is urgently needed to provide information on the glycolytic capacity and immune status of patients while predicting prognosis.
In this study, glycolysis-related genes (GRGs) and immune-related genes (IRGs) were selected as candidate genes, and the optimal candidate gene range was determined using a variety of analytical screening methods. Next, a comprehensive index of glycolysis and immunity (CIGI) model was constructed. CIGI scores are stable and reliable biomarkers in different data sets and different subgroups. Finally, by analyzing the gene expression levels, we showed that CIGI scores reflect the glycolytic capacity and immune status of LUAD patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1695/rc).
Methods
Data source: online database
Ribonucleic acid (RNA)–sequencing gene expression information for all the samples and clinical information for LUAD patients were obtained from The Cancer Genome Atlas (TCGA) database and the Gene Expression Omnibus (GEO) database (GSE31210, GSE41271, and GSE50081). The TCGA dataset was used as training set, and the GEO datasets were used as validation sets. All samples with deficient clinical information such as gender, tumor stages and OSs were excluded from the study. The ENSEMBL IDs in all the sequencing data were converted to gene symbols. When the same probe corresponded to multiple genes, it was excluded, and when multiple probes corresponded to the same gene, the median of the expression values was used for the analysis. GRGs and IRGs were identified in the Molecular Signatures database and the ImmPort data set.
CIGI build and verification
A univariate analysis was performed using the “survival” package in the GRGs and IRGs to select the independent prognostic predictors. The CIGI model was constructed using a least absolute selection and shrinkage operator (LASSO) analysis and Cox proportional hazards regression models. The risk value of each patient was calculated, and the LUAD patients were then divided into two groups according to the optimized risk value. The difference in survival between the two groups was examined via a Kaplan-Meier (K-M) survival analysis. The predicted values of the CIGI were analyzed by time-dependent receiver operator characteristic (ROC) curves. Multiple factors that may affect the prognosis, such as gender, T (primary tumor) stage, N (lymph node) stage, M (metastasis) stage, and tumor stage were included in COX regression analysis to identify independent prognostic factors. Based on the results of multivariate Cox regression analysis, a nomogram model was established to predict the prognosis. The calibration curves and the ROC curves were used to validate the nomogram model.
Analysis of potential regulatory pathways and immune infiltration
The pathway scores of each sample and the infiltration levels of the immune cells were analyzed by a single-sample gene set enrichment analysis (ssGSEA). The relationship between the CIGI and potential regulatory pathways was analyzed based on the scores. Genetic markers for each immune cell were determined from previous research (12).
IHC analysis
The LUAD samples and matched non-tumorous tissues were obtained from 80 LUAD patients in the First Affiliated Hospital of Anhui Medical University. The tissues were fixed in 10% formalin, embedded in paraffin, and processed as 4-µm continuous sections. Immunohistochemistry (IHC) staining was performed according to the manufacturers’ instructions (UltraSensitiveTM SP; MXB, China). The following antibodies were used: fibroblast growth factor (FGF2; Abcam, ab92337), hyaluronan-mediated motor receptor (HMMR; Proteintech, 15820-1-AP), and nuclear receptor 0B2 (NR0B2; Abcam, ab96605). Each sample was independently assessed by 2 pathologists and scored using a semi-quantitative scoring system with histoscores ranging from 0 (minimum) to 300 (maximum). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the First Affiliated Hospital of Anhui Medical University (No. PJ2018-16-15) and individual consent for this retrospective analysis was waived.
Statistical analysis
Multiple factors that may affect the prognosis were included in univariate COX and multivariate COX regression analysis, and independent prognostic factors were selected according to P<0.05. A K-M analysis was conducted to analyze the differences in OS. Statistical comparisons between two groups were performed using the Student’s 2-tailed t-test. A P value <0.05 was considered statistically significant. The P value was two sided. The threshold AUC value is 0.5.
Results
Construction of the LUAD CIGI model
We first performed an analysis using TCGA-LUAD data set. Differentially expressed genes between the LUAD tissues and adjacent paracancerous tissues were screened using the following standard: a log 2-fold change (FC) >1 and P<0.05 (Figure 1A). In total, 1,491 differentially expressed genes (of which 574 were upregulated and 917 were downregulated) were identified. After intersecting these differentially expressed genes with the IRGs/GRGs, 144 genes were identified (Figure 1B). We further analyzed the association between these 144 genes and OS and disease-free survival (DFS), and found that 18 genes were significantly associated with OS and DFS (Figure 1C-1E). To identify more precise candidate genes, we performed a LASSO regression analysis and cross-validated the regression coefficients and ultimately identified 9 candidate genes (Figure 1F,1G). To examine the relationship between these 9 candidate genes and survival time and to optimize the final prediction model, we identified a set of 3 genes (i.e., FGF2, HMMR, and NR0B2) using a Cox proportional hazards regression model. The analysis result was as follows: CIGI = (0.269× FGF2 expression level) + (0.177× HMMR expression level) + (–0.076× NR0B2 expression level).
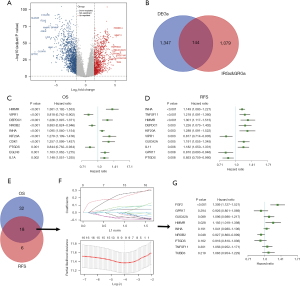
Prognostic value of CIGI score in TCGA-LUAD data set
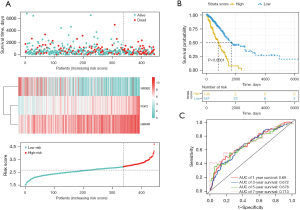
We calculated the risk score according to the CIGI model. The survival time distribution, expression levels of the 3 genes, and risk score distribution of the LUAD patients are shown in Figure 2A. The risk values for all patients were calculated, and the best cut-off value was selected. Patients were assigned to two groups according to their scores. The K-M survival analysis showed that patients in the low-risk group had significantly longer OS than those in the high-risk group (Figure 2B; P<0.0001). A ROC curve analysis was conducted to verify the predictive ability of the model, and the area under the curve (AUC) values for 1-, 3-, and 5-year survival were 0.69, 0.672, and 0.678, respectively (Figure 2C).
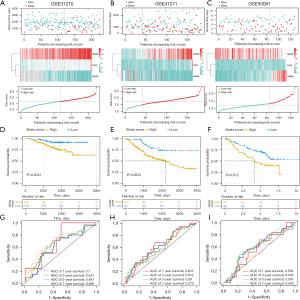
CIGI validation in GEO data sets
To validate the prognostic value of CIGI, we selected 3 external data sets. The reliability of the CIGI predictions was tested on 3 data sets; that is, GSE31210, GSE41271 and GSE50081. The survival times, expression levels of the 3 genes, and risk score distributions for the 3 cohorts are shown in Figure 3A-3C. The K-M survival analysis showed that in all 3 cohorts, patients with low scores had longer OS (Figure 3D-3F). A ROC curve analysis was used to verify the predictive power of the predictive model. The AUC values for 1-, 3-, 5-, and 7-year survival in GSE31210 were 0.7, 0.612, 0.647, and 0.695, respectively (Figure 3G). The AUC values for 1-, 3-, 5-, and 7-year survival in GSE41271 were 0.616, 0.612, 0.59, and 0.579, respectively (Figure 3H). The AUC values for 1-, 3-, 5-, and 7-year survival in GSE50081 were 0.558, 0.595, 0.591, and 0.543, respectively (Figure 3I).
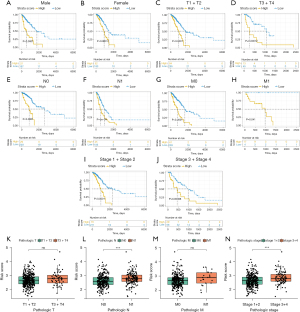
Prognostic performance of the CIGI in different subgroups
To more comprehensively and more closely evaluate the clinical predictive value of the CIGI, patients with LUAD were divided into different subgroups according to their clinical characteristics, and a K-M survival analysis was performed. The results showed that OS was longer in the low-score group in subgroups of LUAD patients by gender, T (primary tumor) stage, N (lymph node) stage, M (metastasis) stage, and tumor stage (Figure 4A-4J). A distribution analysis of the CIGI scores was also performed in the LUAD subgroups of patients with different T, N, M, and pathological stages, and the results showed that patients in the high CIGI group had later N and tumor stages (Figure 4K-4N). In conclusion, CIGI effectively predicted the prognosis of different subgroups of patients, which proves the accuracy and reliability of the CIGI.
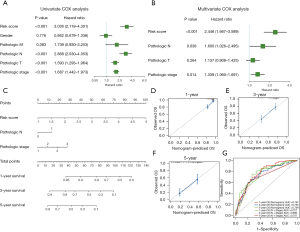
The Cox analysis showed that CIGI is an independent prognostic factor of LUAD
First, a univariate Cox analysis was performed to explore the correlations between several candidate factors and prognosis. The candidate factors included the CIGI and 5 common clinical features. The results showed that 4 factors (i.e., CIGI, N stage, T stage, and tumor stage) were statistically significant for the prognosis of LUAD patients (Figure 5A). Next, a further multivariate Cox analysis was performed on the above 4 factors. The results showed that 3 of the 4 factors (i.e., CIGI, N stage, and tumor stage) were statistically significant independent factors in the prognosis of LUAD patients (Figure 5B). In conclusion, the CIGI value is an independent prognostic factor in patients with LUAD. To further examine the prognostic predictive function of the CIGI values for LUAD patients, we incorporated all the independent prognostic factors and constructed a nomogram model to increase its predictive power (Figure 5C). The calibration curves showing the 1-, 3-, and 5-year calibration points were in good agreement with the standard curve, indicating that the model exhibited a valid predictive performance (Figures 5D-5F). Further, the ROC analysis suggested that the nomogram model was more predictive of OS than a single clinicopathological feature (Figure 5G).
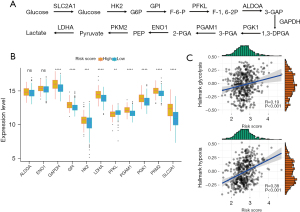
The CIGI is able to differentiate glycolytic status in LUAD patients
To further evaluate the relationship between CIGI and glycolysis, we identified multiple genes that encode cellular glycolysis regulation based on previous reports (Figure 6A) (13). We then analyzed the messenger RNA expression levels of the 11 genes in patients with low and high CIGI scores using TCGA database. According to TCGA data set, except for ALDOA (aldolase A), ENO1 (α-enolase), and PFKL (phosphofructokinase-1 liver type), the other GRGs showed higher expression levels in the high CIGI group (Figure 6B). In addition, the ssGSEA scores of the “HALLMARK GLYCOLYSIS” and “HALLMARK HYPOXIA” pathways were also generated based on the RNA expression profiles of TCGA data set, and the correlation analysis suggested that both the glycolytic pathway score and the hypoxia pathway score were positively correlated with the CIGI (Figure 6C). From the above results, we concluded that the expression levels of these genes in patients in the high CIGI group were higher than those patients in low CIGI group, suggesting that patients in the high CIGI group may have stronger glycolytic potential.
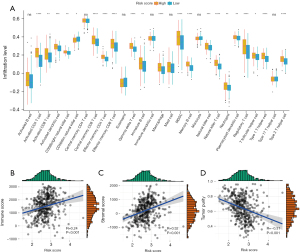
CIGI can differentiate among the different immune statuses of LUAD patients
To explore the link between immune status and the CIGI in LUAD patients, we conducted a ssGSEA to determine the correlations between the risk scores and immune cells and the infiltration levels of the immune cells. The results indicated that in patients with high CIGI, 21 immune cells showed high infiltration levels (Figure 7A). To further evaluate the relationship between the CIGI and the tumor immune microenvironment, the status of the tumor immune microenvironment was evaluated based on the stromal score, immune score, and tumor purity. As Figure 7B-7D shows, the CIGI scores were weakly positively correlated with immune scores (R=0.24, P<0.001) and stromal scores (R=0.32, P<0.01) but were weakly negatively correlated with tumor purity (R=–0.31, P<0.001). The above results suggest that patients with higher CIGI scores had relatively higher levels of immune cell infiltration, which also suggests that the immune level of such patients may be stronger.
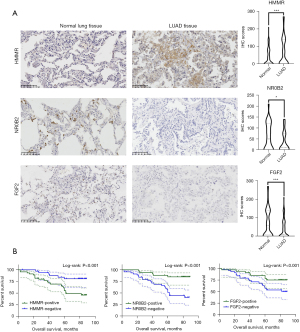
The protein expression of FGF2, HMMR, and NR0B2 in LUAD patients
To further verify the expression of FGF2, HMMR, and NR0B2 in the LUAD patients, we conducted IHC experiments in 80 LUAD patients’ tumor tissues and adjacent tissues to detect the protein expression levels of the above 3 genes. The results showed that HMMR was highly expressed in the tumor tissues but was lowly expressed in adjacent tissues. Conversely, NR0B2 and FGF2 were highly expressed in the adjacent tissues, but were lowly expressed in the tumor tissues (Figure 8A). We then performed a K-M survival analysis of the patients. The results showed that the patients with high expression of HMMR had a longer OS period, while those with a high expression of NR0B2 and FGF2 had a shorter OS period (Figure 8B). This was consistent with our previous data analysis results.
Discussion
LUAD is a malignant tumor with high morbidity and mortality. Despite multiple treatment options, the prognosis of patients with LUAD, especially those with advanced LUAD, remains poor. Thus, a more detailed classification system for LUAD patients is urgently needed to better predict patient prognosis and provide evidence for more precise treatment. With the development of bioinformatics and sequencing technologies, studies have built predictive models based on immunity and glycolysis, but most models contain only 1 biological process. This study established the first model that combines immunity and glycolysis factors for LUAD.
In this study, we constructed a CIGI model using data from public databases. As shown in different data sets and patient subgroups, the CIGI model had excellent prognostic ability in LUAD patients and the CIGI score could be a key independent prognostic factor in LUCD patients, and thus has great potential for clinical application. Further, we demonstrated that CIGI is associated with glycolytic capacity and immune status. Our findings may also lead to a new research direction for the subsequent elucidation of the driving mechanism of LUAD.
In recent years, several studies have shown that altered metabolic processes in the tumor microenvironment inhibit some immune cell infiltration and other anti-tumor immune processes by producing immunosuppressive metabolites (10,11). Interestingly, some metabolic processes underlie cancer and immune cell responses, while another direct consequence of changes in tumor metabolism is the activation of immunosuppressive pathways. In addition, changes in immune status can also affect cellular metabolism. For example, blocking immune checkpoints can inhibit glycolysis in tumor cells, and enable T-cell glycolysis and cytokine production (14). Thus, an in-depth understanding of tumor metabolism and immune status could enable doctors to selectively modulate related functions while providing an accurate prognostic prediction and guidance for subsequent treatment.
In this study, a prognostic model combining glycolysis and immunity factors was constructed, which reflects the changes in these 2 characteristics of tumors at the same time. Notably, patients with higher CIGI scores had stronger glycolysis, a poorer prognosis, and a later stage of LUAD. However, patients with higher CIGI scores also had stronger immune responses, indicating that the immune responses of these patients did not produce stronger tumor suppressor effects, which suggests that these patients may have immune escape.
In recent years, many researchers have developed new prediction models. In 2020, Luo et al. proposed prognosis models based on IRGs (15,16). However, these models only target IRGs, and do not include metabolic processes, such as glycolysis. In the same year, Zhao et al. also developed a whole genome prediction model; however, this model is aimed at the whole genome, and does not reflect the relationship between patients’ IRGs and GRGs and prognosis (17). Unlike previously established models, our model was the first to combines IRGs and GRGs, and thus faithfully reflects the relationship between these two physiological processes and patient prognosis. Additionally, the CIGI score also indicates the immune infiltration level and glycolysis ability of the patient to some extent. In addition, our model does not include too many genes, which is beneficial for clinical application and detection. The expression of target genes has also been tested in the tissue section verification of patients. Thus, the model proposed in this study is innovative and of great significance for clinical treatment and further research.
The CIGI model we constructed includes the following 3 genes: FGF2, HMMR and NR0B2. FGF2 is an important growth factor necessary for the construction of lung tissue. In certain contexts, FGF2 triggers the abnormally high expression of VEGF (vascular endothelial-derived growth factor) and FGF-2, which are the main drivers of abnormal angiogenesis and lung cancer development, in lung tissue structures by activating the inflammatory cascade (18). FGFR2 (fibroblast growth factor receptor 2)-CCAR2 (cell cycle and apoptosis regulator 2) and other fusions and gain-of-function mutations in FGFR2 have been observed in lung cancer (19-21), which suggests that aberrant FGFR signaling induces lung cancer. In addition, it has also been reported that abnormal FGFR signaling may promote tumor cell proliferation, survival, invasion, and metastasis, involving multiple stages of tumor development.
HMMR was originally identified as a soluble protein that binds to hyaluronic acid with diverse cellular functions. It primarily plays a role in the repair of tissue damage (22,23). A previous study showed that HMMR participates in the formation of microtubule spindles and activates the signaling pathways that enhance cell migration, thereby promoting cell cycle progression (24). A 2021 report noted an inverse correlation between HMMR expression levels and OS in patients with LUAD (25). HMMR has also been proposed as a prognostic marker for early stage non-small cell lung cancer (26).
NR0B2 is an orphan nuclear receptor. NR0B2 expression is lacking in human cancer samples; 2 reports have noted that NR0B2 is downregulated in liver cancer (27) and renal cancer (28). NR0B2 has been reported to suppress inflammation and innate immunity to hepatocyte injury (29-32). Given that inflammatory responses and immune cells have distinct functions in immune escape and anti-tumor immunity (30,33), the clinical significance of NR0B2 expression in relation to tumor immunity requires more research. This study is the first to report that the expression of the above 3 genes in LUAD is related to glycolysis and immunity. Our results suggest that drugs related to FGF2, HMMR, and NR0B2 may become new options for lung cancer-targeted therapy.
Our findings show that the CIGI model is very accurate as a prognostic tool for LUAD patients; however, this study had some limitations. First, all the samples used in our study comprised retrospective cases obtained from public databases, so studies with prospective samples need to be conducted in the future to validate our results obtained. Second, the focus of our study was on the prognostic value of CIGI. The underlying mechanisms behind the prognostic predictive value of FGF2, HMMR, and NR0B2 in CIGI need to be further investigated by more experiments both in vivo and in vitro.
Conclusions
After a series of analyses and verifications, our research showed that the prognosis prediction model established using the expression levels of FGF2, HMMR, and NR0B2, the 3 GRGs and IRGs, accurately predicts the prognosis of LUAD patients. This model also predicts the corresponding glycolysis ability and immune infiltration level of patients.
Acknowledgments
Funding: This work was supported by grants from the Provincial Natural Science Foundation of Anhui (No. 1808085QH232) and the Youth Development Program of The First Affiliated Hospital of Anhui Medical University (No. 2018kj33).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1695/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1695/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1695/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the First Affiliated Hospital of Anhui Medical University (No. PJ2018-16-15) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Chi A, He X, Hou L, et al. Classification of Non-Small Cell Lung Cancer's Tumor Immune Micro-Environment and Strategies to Augment Its Response to Immune Checkpoint Blockade. Cancers (Basel) 2021;13:2924. [Crossref] [PubMed]
- Xia L, Oyang L, Lin J, et al. The cancer metabolic reprogramming and immune response. Mol Cancer 2021;20:28. [Crossref] [PubMed]
- Wu Z, Lee YF, Yeo XH, et al. Shifting the Gears of Metabolic Plasticity to Drive Cell State Transitions in Cancer. Cancers (Basel) 2021;13:1316. [Crossref] [PubMed]
- Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science 2020;368:eaaw5473. [Crossref] [PubMed]
- Renner K, Singer K, Koehl GE, et al. Metabolic Hallmarks of Tumor and Immune Cells in the Tumor Microenvironment. Front Immunol 2017;8:248. [Crossref] [PubMed]
- Callao V, Montoya E. Toxohormone-like factor from microorganisms with impaired respiration. Science 1961;134:2041-2. [Crossref] [PubMed]
- Kim J, DeBerardinis RJ. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab 2019;30:434-46. [Crossref] [PubMed]
- Xu K, Yin N, Peng M, et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 2021;371:405-10. [Crossref] [PubMed]
- Watson MJ, Vignali PDA, Mullett SJ, et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 2021;591:645-51. [Crossref] [PubMed]
- Kaymak I, Williams KS, Cantor JR, et al. Immunometabolic Interplay in the Tumor Microenvironment. Cancer Cell 2021;39:28-37. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Li L, Liang Y, Kang L, et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell 2018;33:368-385.e7. [Crossref] [PubMed]
- Chang CH, Qiu J, O'Sullivan D, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015;162:1229-41. [Crossref] [PubMed]
- Luo C, Lei M, Zhang Y, et al. Systematic construction and validation of an immune prognostic model for lung adenocarcinoma. J Cell Mol Med 2020;24:1233-44. [Crossref] [PubMed]
- Sun S, Guo W, Wang Z, et al. Development and validation of an immune-related prognostic signature in lung adenocarcinoma. Cancer Med 2020;9:5960-75. [Crossref] [PubMed]
- Zhao J, Guo C, Ma Z, et al. Identification of a novel gene expression signature associated with overall survival in patients with lung adenocarcinoma: A comprehensive analysis based on TCGA and GEO databases. Lung Cancer 2020;149:90-6. [Crossref] [PubMed]
- Laddha AP, Kulkarni YA. VEGF and FGF-2: Promising targets for the treatment of respiratory disorders. Respir Med 2019;156:33-46. [Crossref] [PubMed]
- Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-19. [Crossref] [PubMed]
- Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636-47. [Crossref] [PubMed]
- Tanizaki J, Ercan D, Capelletti M, et al. Identification of Oncogenic and Drug-Sensitizing Mutations in the Extracellular Domain of FGFR2. Cancer Res 2015;75:3139-46. [Crossref] [PubMed]
- He Z, Mei L, Connell M, et al. Hyaluronan Mediated Motility Receptor (HMMR) Encodes an Evolutionarily Conserved Homeostasis, Mitosis, and Meiosis Regulator Rather than a Hyaluronan Receptor. Cells 2020;9:819. [Crossref] [PubMed]
- Tolg C, Hamilton SR, Nakrieko KA, et al. Rhamm-/- fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J Cell Biol 2006;175:1017-28. [Crossref] [PubMed]
- Connell M, Chen H, Jiang J, et al. HMMR acts in the PLK1-dependent spindle positioning pathway and supports neural development. Elife 2017;6:28672. [Crossref] [PubMed]
- Li X, Zuo H, Zhang L, et al. Validating HMMR Expression and Its Prognostic Significance in Lung Adenocarcinoma Based on Data Mining and Bioinformatics Methods. Front Oncol 2021;11:720302. [Crossref] [PubMed]
- He R, Zuo S. A Robust 8-Gene Prognostic Signature for Early-Stage Non-small Cell Lung Cancer. Front Oncol 2019;9:693. [Crossref] [PubMed]
- Zhu R, Tu Y, Chang J, et al. The Orphan Nuclear Receptor Gene NR0B2 Is a Favorite Prognosis Factor Modulated by Multiple Cellular Signal Pathways in Human Liver Cancers. Front Oncol 2021;11:691199. [Crossref] [PubMed]
- Prestin K, Olbert M, Hussner J, et al. Modulation of expression of the nuclear receptor NR0B2 (small heterodimer partner 1) and its impact on proliferation of renal carcinoma cells. Onco Targets Ther 2016;9:4867-78. [Crossref] [PubMed]
- Zou A, Magee N, Deng F, et al. Hepatocyte nuclear receptor SHP suppresses inflammation and fibrosis in a mouse model of nonalcoholic steatohepatitis. J Biol Chem 2018;293:8656-71. [Crossref] [PubMed]
- Yuk JM, Jin HS, Jo EK. Small Heterodimer Partner and Innate Immune Regulation. Endocrinol Metab (Seoul) 2016;31:17-24. [Crossref] [PubMed]
- Go MJ, Noh JR, Hwang JH, et al. Small heterodimer partner deficiency exacerbates binge drinking-induced liver injury via modulation of natural killer T cell and neutrophil infiltration. Mol Med Rep 2018;17:4989-98. [Crossref] [PubMed]
- Zhou H, Wang H, Ni M, et al. Glycogen synthase kinase 3β promotes liver innate immune activation by restraining AMP-activated protein kinase activation. J Hepatol 2018;69:99-109. [Crossref] [PubMed]
- Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol 2020;17:807-21. [Crossref] [PubMed]


