
High SERPINH1 expression predicts poor prognosis in lung adenocarcinoma
Highlight box
Key findings
• High SERPINH1 expression predicts poor prognosis in lung adenocarcinoma.
What is known and what is new?
• Previous studies have shown that SERPINH1 exhibits high expression in a variety of tumors. The aberrant expression of this gene is closely linked to tumor growth, invasion, and metastasis. To date, no studies had been conducted on the expression of SERPINH1 in LUAD and its potential prognostic relevance.
• we comprehensively assessed the prognosis of SERPINH1 and LUAD and their relationship to immunotherapy and chemotherapy and obtained good validation results from several perspectives.
What is the implication, and what should change now?
• Our findings provide a new approach and strategy for the clinical treatment of LUAD patients. The in-vivo and in-vitro experimental validation of SERPINH1 is needed.
Introduction
Lung cancer is the leading cause of cancer-related deaths and one of the most serious malignancies threatening human health and life (1). The incidence and mortality rates of lung cancer have risen dramatically in recent years (2), with both men and women experiencing the 2nd highest incidence of any malignancy (3). At the time of diagnosis, the majority of lung malignancies are already either locally progressed or metastatic. There are two types of lung cancer (i.e., small cell lung cancer and non-small cell lung cancer), of which non-small cell lung cancer comprises about 85% of all lung cancer cases (4). Lung adenocarcinoma (LUAD) accounts for 40% of lung cancers (5,6) and 55% of non-small cell lung cancers. For early-stage non-small cell lung cancer, the main treatment modality is surgical resection (7). For advanced-stage non-small cell lung cancer, treatment options include chemotherapy (8), molecular targeted therapy (9), and immunotherapy (10). The etiology of LUAD development is still unclear, and it is crucial to explore new prognostic biomarkers and therapeutic targets.
As one of the most frequently amplified areas of human malignancies, Serpine Protease Inhibitorclade H1 (SERPINH1) is on chromosome 11q13.5 and encodes the protein heat shock protein 47 (HSP47), which is an important molecular chaperone required for the proper folding and secretion of various types of collagen (11). SERPINH1 has been identified as a possible biomarker for several cancer types (12-15), and studies have shown that the aberrant expression of this gene is closely linked to tumor growth, invasion, and metastasis (16,17).
Collagen is the most abundant protein in the body and a major component of the extracellular matrix (ECM) (18). Extracellular matrix changes have been found to be strongly associated with cancer development and progression (19), particularly collagen remodeling (20) and collagen deposition (21). A study has confirmed that SERPINH1 may promote cancer growth and invasion by regulating the extracellular matrix (22). First, inhibition of SERPINH1 expression reduces collagen deposition (23). Secondly, Epithelial-Mesenchymal Transition (EMT) can promote tumor invasion and metastasis (24,25). SERPINH1 can regulate EMT by modulating the Wnt/β-catenin signaling pathway to affect tumor development (26).
Although (27) and others have made a preliminary exploration of SERPINH1 expression on lung adenocarcinoma prognosis, our study is innovative in the following ways. Firstly, there are more reasonable and innovative sources of data for our study. We have downloaded LUAD data from The Cancer Genome Atlas (TCGA) database, standardized the data and the results are reasonable and reliable. Secondly, we analyzed the prognosis of high and low SERPINH1 expression and lung adenocarcinoma by collecting clinical data samples from Affiliated Hospital of Nantong University. In addition, for exploring the functional study of SERPINH1 in lung adenocarcinoma, we first identified the genes co-expressed with SERPINH1 in lung adenocarcinoma by performing enrichment analysis of GO and KEGG pathways on the co-expressed genes, which resulted in more reliable analysis. Our study is the first to collect clinical data for the analysis of SERPINH1 and lung adenocarcinoma prognosis. This is the first systematic assessment of SERPINH1 expression in relation to prognosis, associated pathways, tumor mutation burden (TMB) and immune infiltration, immune checkpoint inhibitors, and antitumor drug sensitivity in lung adenocarcinoma.
The tumor microenvironment can provide a specific environment and plays a non-negligible role in tumor development (28,29). The level of immune infiltration in the tumor microenvironment can promote or suppress the biological behavior of tumors (30-33). Previous studies have shown that SERPINH1 is associated with tumor immune cell infiltration and may have an impact on tumor prognosis (13,27). Therefore, we further explored the correlation of SERPINH1 and immune infiltration in lung adenocarcinoma.
This study examined SERPINH1 and LUAD prognosis and related pathway mechanisms and found considerable variations in the prognosis, TMB, immune infiltration, and treatment sensitivity of patients with high and low expression of SERPINH1. Our findings suggest that SERPINH1 may be crucial in the growth of LUAD and could be used as a cutting-edge and precise tool for clinical diagnosis and individualized care. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1518/rc).
Methods
Collection of LUAD data
In this study, 598 transcriptomic samples (539 tumor tissue samples and 59 normal tissue samples) and 522 clinically relevant LUAD samples were downloaded from TCGA database (https://portal.gdc.cancer.gov/). After eliminating samples with incomplete clinical data and missing survival times, 503 samples remained for further investigation.
Collection of LUAD tissue microarray (TMA) samples
TMA samples were obtained from Affiliated Hospital of Nantong University (NTU). All the samples were collected from patients who had undergone lung cancer resection without other primary sites from 2009 to 2010. These TMA samples had complete clinicopathological data on lung adenocarcinoma and 10-year follow-up information. In total, 140 LUAD tissue samples and 15 normal tissue samples were collected. Information from the medical records of the 140 LUAD patients was collected, including age, sex, pathological diagnosis, tumor diameter, lymph node status (N), clinical stage, pathological grade, survival status, survival time, and smoking history. Informed consent was obtained from the patients to use their human tissue for this study. The study was approved by the Clinical Research Ethics Committee of Affiliated Hospital of Nantong University (No. 2022-L094). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Analysis of differential expression of SERPINH1 in different tumors
To explore the expression of SERPINH1 in different tumors, we analyzed the results using the hiplot (https://hiplot-academic.com) online website.
IHC staining and scoring
Surgical specimens that had been formalin-fixed and paraffin-embedded were used to create the TMAs. Tissue blocks were cut to a thickness of 4 µm and prepared as TMAs on slides. Immunohistochemistry investigations were then performed to evaluate SERPINH1 expression. This was done as follows: after dewaxing, endogenous horseradish peroxidase (HRP) was inactivated with hydrogen peroxide, and the tissue was subjected to antigen repair using sodium citrate antigen repair solution at high temperature and pressure. The tissue was then closed with phosphate-buffered solution (PBS) containing 5% Blocking Buffer (BSA), and 0.3% TritonX-100, for 2 h at room temperature. After rinsing, a rabbit anti-SERPINH1 antibody (1:200, Abcepta) was diluted with antibody diluent and the tissue was incubated with this antibody overnight. The next day, PBS was washed the tissue 3 times, and then murine general secondary antibody HRP (k8002, DAKO, Denmark) was dropwise incubated at 37 ℃ for 120 min. Finally, diaminobenzidine color development, hematoxylin-stained nuclei, gradient dehydration, and neutral gum sealing were carried out. Each specimen was independently re-evaluated by 2 pathologists following the Tumor Node Metastasis (TNM) categorization criteria used by the International Union Against Cancer. The staining intensity levels were described as follows: negative [0], weak [1], moderate [2], and strong [3], while the degree of staining was scored according to the percentage of positive cells: 0 (0%); 1 (1–25%); 2 (26–50%); 3 (51–75%); and 4 (76–100%). Finally, a composite score was calculated by multiplying the intensity and density of staining; a low expression of SERPINH1 was defined as ≤2, and a high expression of SERPINH1 was defined as ≥3.
Survival analysis
We used the “limma” package to analyze the variations in SERPINH1 expression between LUAD cases and normal paracancerous tissue samples using clinical data from the TCGA database. The median value of SERPINH1 expression was used to group samples according to their level of expression, and the survival time and survival status of LUAD patients were merged with SERPINH1 expression. There were 251 samples with high expression and 252 samples with low expression. The 140 LUAD samples collected from Affiliated Hospital of Nantong University were scored according to the intensity and density of staining, with the low expression of SERPINH1 defined as ≤2, and the high expression of SERPINH1 defined as ≥3. The survival analysis was performed using the “survminer R” package and the “survival R” package for the high and low expression groups, and a P value <0.05 indicated a statistically significant difference between the 2 groups.
Comprehensive clinical prognostic analysis
Clinical data downloaded from the TCGA database were used for the analysis. A univariate Cox regression analysis was conducted to initially explore the relationship between SERPINH1 and age, gender, and stage. A multivariate Cox analysis was used to verify whether SERPINH1 could be used as an independent prognostic factor in LUAD. A univariate Cox regression analysis was used to primarily investigate the association between SERPINH1 and age, gender, T stage, N stage, and pathological grade using tissue data from the 140 LUAD cases obtained from Affiliated Hospital of Nantong University. The multivariate Cox analysis was used to confirm whether SERPINH1 could be employed as an independent prognostic risk factor. The “ggplot2” package was used for the visualization process. Using the “complexHeatmap” package, clinical correlation heat maps were generated.
Establishment of nomogram and analysis
Based on the univariate and multivariate Cox analyses, we used SERPINH1, gender, age, and stage to construct a TCGA column nomogram to predict the survival rates of LUAD patients at 1, 3, and 5 years. The NTU group nomograms were constructed using SERPINH1, gender, T stage, and N stage to predict the survival rates of LUAD patients at 1, 3, and 5 years. The nomogram was constructed using the “rms” and drew using the “regplot” packages. We used calibration curves to assess the consistency of the model predictions. The diagnostic value of the nomogram was evaluated using the receiver operating characteristic (ROC) curve, with a larger area under the curve (AUC) representing a higher diagnostic value.
SERPINH1 co-expression and enrichment analysis
First, we used the “limma” package to assess the genes related to SERPINH1. Those with correlation coefficients >0.5 and P values <0.001 were considered significantly associated genes and selected for further study. Further, we chose the top 100 genes related to SERPINH1 for the enrichment analysis. Using the “clusterProfiler” R package, we ran enrichment analyses for the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) based on the associated genes we had identified.
Assessing TMB values and the relationship with survival
The TMB was calculated using single-nucleotide variant data from LUAD patients acquired from the TCGA database. The link between the risk score and the TMB was investigated using the Spearman correlation analysis. To evaluate TMB and the prognostic differences between the high and low-expression groups, a Kaplan-Meier survival curve analysis was used. The “maftools” R package was used to calculate the number of non-synonymous point mutations in somatic cells within each sample.
SERPINH1 correlation analysis with the TME and immune infiltration
The Estimation of Stromal and Immune Cells in Malignant Tumors using the Expression Data (ESTIMATE) algorithm was used to calculate ESTIMATE, immune, and stromal scores to predict the level of infiltrating immune and stromal cells. Next, using box plots to show the outcomes of the CIBERSORT analyses and single-sample gene set enrichment analyses (ssGSEAs), we used the CIBERSORT algorithm to score the common 22 immune cells and the ssGSEA algorithm to evaluate the common 29 immune cell features. For the Wilcoxon test, a P value <0.05 was considered statistically significant.
Correlation analysis of SERPINH1 with immune checkpoints and anti-tumor drugs
We compared the differences between SERPINH1 and common immune checkpoints with the Wilcoxon rank-sum test and correlated the 2 with the Spearman test. The associations between SERPINH1 and 8 popular anti-cancer medications were then examined. The 50% reduction in growth concentration (IC50) of chemotherapeutic agents was calculated using the R package “pRRophetic”. The Wilcoxon rank-sum test was used to compare the variations in IC50 between the high and low-expression groups.
Statistical analysis
All the statistical analyses were conducted, and the graphs were generated using an R programming language (version 4.1.1). The expression of SERPINH1 in the unpaired samples was analyzed using the Wilcoxon rank-sum test. The expression of SERPINH1 in the paired samples was analyzed using the paired samples t-test, and P<0.05 was considered statistically significant. Kaplan-Meier survival analysis and the log-rank test were used to estimate the survival distributions. For various clinical features, the Cox regression analysis evaluated the risk ratios (HR) and 95% confidence intervals (CIs) and determined the independent prognostic markers.
Results
Differential expression analysis of SERPINH1 in LUAD and normal lung tissues and clinicopathological characterization
To investigate the expression of SERPINH1 in different tumors, we performed a differential expression analysis, and the results showed that SERPINH1 was significantly more highly expressed in breast, colon, LUAD, lung squamous cell carcinoma, and gastric adenocarcinoma tissues than normal tissues (see Figure 1A). Next, we looked at the expression of SERPINH1 in lung cancer tissues, and the results showed that SERPINH1 was more highly expressed in LUAD tissues than in paraneoplastic tissues (see Figure 1B). A paired difference analysis revealed that SERPINH1 was abundant in lung cancer samples (see Figure 1C). Subsequently, we performed immunohistochemical (IHC) staining of the normal lung tissues and LUAD tissues collected at Affiliated Hospital of Nantong University, and the results showed that SERPINH1 expression was significantly higher in the LUAD tissues than in the normal lung tissues (see Figure 1D). Based on IHC scores for high and low SERPINH1 expression, we also performed a differential analysis of 15 normal samples and 140 LUAD tissue samples obtained from Affiliated Hospital of Nantong University. The results revealed that SERPINH1 expression differed in normal lung tissues and tumor tissues and was more highly expressed in tumor tissues (see Figure 1E).
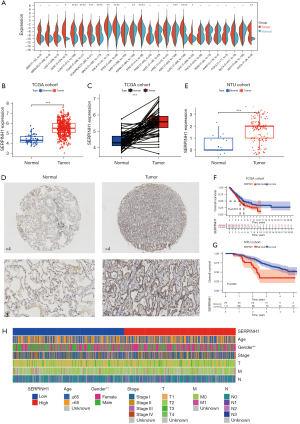
Additionally, we divided the 503 LUADs into a high-expression group (n=251) and a low-expression group (n=252) based on the median value of SERPINH1 expression. The 140 LUAD samples collected from Affiliated Hospital of Nantong University were scored according to the intensity and density of staining, with low expression of SERPINH1 defined as ≤2 and high expression of SERPINH1 defined as ≥3. According to both Kaplan-Meier survival curves, the low-expression group had a greater survival advantage than the high-expression group (see Figure 1F, P=0.018; see Figure 1G, P=0.002). The heat map shows the differences in the clinically relevant characteristics (including age, gender, stage, T stage, N stage, and M stage) between the high and low SERPINH1 expression groups (see Figure 1H). In summary, SERPINH1 differed significantly between the normal lung tissue and LUAD tissue and may play a vital role in tumor development.
Construction and validation of a nomogram
To further explore the relationship between SERPINH1 and overall survival (OS), we performed univariate and multivariate Cox analyses on the TCGA cohort. The univariate Cox analysis showed that SERPINH1 and stage were significantly associated with OS (P<0.05), while the multivariate Cox analysis showed that SERPINH1 and stage were independent prognostic factors for LUAD (P<0.05) (see Figure 2A,2B). The survival of LUAD patients was then predicted using a nomogram based on age, gender, stage, and SERPINH1 expression at 1, 3, and 5 years (see Figure 2C). The calibration curves showed that the actual and expected OS values were quite similar (see Figure 2D). The ROC curves were then used to evaluate the accuracy of the model at 1, 3, and 5 years. According to a nomogram, the AUCs for 1-, 3-, and 5-year OS were 0.718, 0.712, and 0.702, respectively (see Figure 2E).
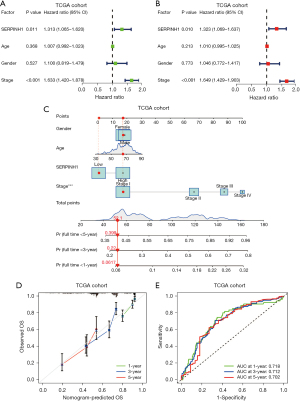
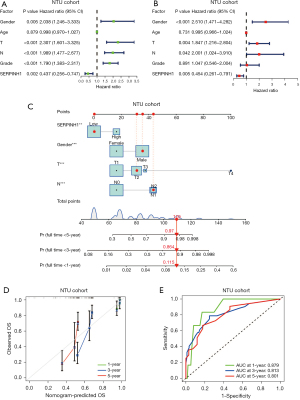
Similarly, we performed univariate and multivariate Cox analyses on the clinical data collected from 140 LUAD cases at Affiliated Hospital of Nantong University. According to the results of the univariate Cox analysis. SERPINH1, T stage, N stage, and pathological grade were all significantly linked to OS (P<0.05) (see Figure 3A). The multivariate Cox analysis showed that SERPINH1, gender, T stage, and N stage were independent prognostic factors for LUAD (P<0.05) (see Figure 3B). Next, using the factors of gender, T stage, N stage, and the expression of SERPINH1, we created a nomogram to predict the survival of LUAD patients at 1, 3, and 5 years (see Figure 3C). The calibration curves showed a general fit between the actual OS values and the predicted OS values (see Figure 3D). The accuracy of the model was then assessed using ROC curves at 1, 3, and 5 years. The areas under the ROC curves at 1, 3, and 5 years were 0.879, 0.813, and 0.801, respectively (see Figure 3E).
Co-expression analysis and functional enrichment analysis of SERPINH1
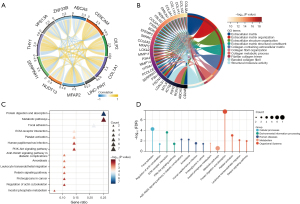
We conducted co-expression and functional enrichment analyses of the SERPINH1 gene to better investigate the biological roles of this gene. Figure 4A shows the top 5 co-expressed genes positively and negatively associated with SERPINH1. The extracellular matrix (ECM) and collagen fibrils were substantially enriched for GO terms among the top 100 co-expressed genes that were most closely related to one another (see Figure 4B). The KEGG enrichment analysis showed that SERPINH1 was mainly associated with metabolic pathways, cell proliferation, and migration (see Figure 4C,4D).
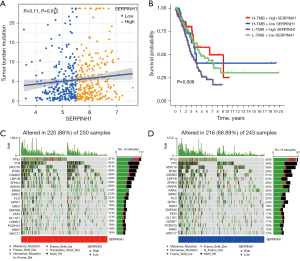
Correlation analysis between SERPINH1 and the TMB
The TMB is positively associated with programmed death-ligand 1 expression, and higher TMB levels are more effective for immunotherapy (34-37). The TMB was found to be positively correlated with clinically lasting benefits and progression-free survival in non-small cell lung cancer immunotherapy (38). The TMB is a biomarker that predicts tumor behavior and treatment response (38). Thus, we combined SERPINH1 with the TMB to explore the potential interactions between the two factors. The Spearman analysis showed that SERPINH1 gene expression and tumor mutations were positively correlated (see Figure 5A). Patients with a high TMB and high SERPINH1 expression had a greater survival advantage according to the stratified survival analysis, while patients with a low TMB and high SERPINH1 expression had worse survival (see Figure 5B). The “maftool” package was then used to determine the somatic mutation rates, and a waterfall plot was used to display the top 20 mutation driver genes. In the SERPINH1 high expression group, tumor protein p53 (TP53) had the highest mutation frequency (47%), followed by Titin (TTN) (43%) and Mucin 16 (MUC16) (41%). However, in the SERPINH1 low expression group, TP53 (40%) had the highest mutation rate among the driver genes (see Figure 5C,5D).
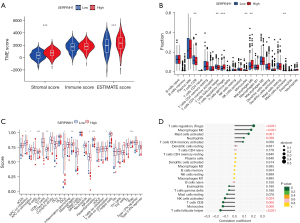
Correlation analysis of SERPINH1 and immune infiltration
Previous research has shown that the immunological microenvironment is essential for the development of tumors (39). Immunological cell infiltration in the immune microenvironment either inhibits or promotes the growth of tumors (40-43). Thus, we further explored the role of SERPINH1 in immune infiltration. We graded the immune and stromal components of the tumor using the ESTIMATE algorithm to forecast its stromal and immune cell composition. The results showed that the SERPINH1 high expression group had higher stromal scores and estimated scores (see Figure 6A). Additionally, we scored 22 immune cells using the CIBERSORT algorithm and discovered that the SERPINH1 low expression group had a larger amount of immunological infiltration in immune cells, such as the cluster of differentiation (CD)8+ T cells, follicular helper T cells, and monocytes. Conversely, regulatory T cells, M0 macrophages, and activated mast cells had higher immune scores in the SERPINH1 high expression group than the SERPINH1 low expression group (see Figure 6B). Next, using the ssGSEA algorithm, we evaluated 29 common immune cells and immunological functions. We found that the SERPINH1 high-expression group had higher expression levels of immunological functions, such as antigen presentation, chemokines, and immune checkpoints, than the low-expression group (see Figure 6C). Finally, we assessed the correlation between 22 immune cells and SERPINH1 using a Pearson correlation analysis, the results of which are shown in Figure 6D. In conclusion, we discovered that various SERPINH1 expressions displayed various immune infiltration traits. SERPINH1 most likely contributes significantly to immune infiltration and TME.
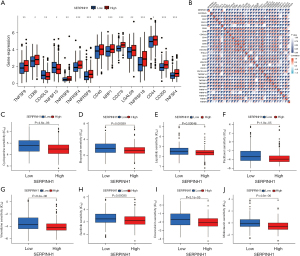
Analysis of SERPINH1 and immune checkpoints and drug correlations
The approaches used to treat malignancies have evolved significantly in recent years, especially with the introduction of immune checkpoint blockade medication, which has altered how many cancers were previously treated. Immune checkpoint blockade therapy, which is mainly used to activate the immune system of the body by targeting cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), or Programmed cell death 1 ligand 1 (PD-L1), induces T cells to kill tumor cells, thus achieving a therapeutic effect (44). The standard of care for non-small cell lung cancer now includes immune checkpoint blockade medication, which greatly increases survival (45). We first analyzed popular immune checkpoint genes to assess the potential propensity of SERPINH1 to predict treatment in LUAD patients. The results showed that TNFSF9, CD86, TNFRSF8, TNFRSF4, TNFRSF9, CD40, NRP1, CD276, LGALS9, TNFRSF18, CD44, CD200, and TNFSF4 were more highly expressed in the SERPINH1 high expression group than the SERPINH1 low expression group, while CD40LG and TNFSF15 were more highly expressed in the SERPINH1 low expression group (see Figure 7A). Subsequently, the Pearson correlation analysis showed that most of the immune checkpoints were positively correlated with SERPINH1 (see Figure 7B). This suggests that patients in the high SERPINH1 expression group will have better treatment outcomes when receiving immunotherapy. We also analyzed the common clinical anti-tumor drugs and the results showed that the 8 common anti-tumor drugs (i.e., cyclopamine, etoposide, lapatinib, paclitaxel, vinorelbine, sunitinib, doxorubicin, and midostaurin) were more effective in the IC50 of the SERPINH1 high expression group was significantly lower than that of the low expression group (see Figure 7C-7J). This suggests that anti-tumor therapy was more efficacious in the SERPINH1 high-expression group than the SERPINH1 low-expression group. In brief, the immunotherapy and anti-tumor treatment produced better results for the SERPINH1 high-expression group than the SERPINH1 low-expression group, and the study of SERPINH1 has the potential to provide a new approach and strategy for the clinical treatment of LUAD patients.
Discussion
LUAD is a type of non-small cell lung cancer and accounts for approximately 40% of lung cancers (6). The development of lung cancer is a progressive process that involves many steps and factors. The main causes of LUAD include smoking, air pollution, and chronic lung conditions. Previous studies have shown that lung cancer has a high mortality rate and a poor survival rate, is one of the malignancies with the worst prognosis (46), and is expected to become the primary burden of human cancer by 2050 (47). Thus, efficient biomarkers for the early detection and prognostication of lung cancer urgently need to be found.
SERPINH1 encodes the HSP47 protein that is required for the proper folding and secretion of collagen (11). Previous studies have shown that SERPINH1 exhibits high expression in a variety of tumors, including cervical cancer (16), colorectal cancer (48), gastric cancer (26), and breast cancer (22). In cervical cancer, it was found that when SERPINH1 was silently expressed in cervical cancer cells, the invasion and migration ability of the cervical cancer cells was significantly inhibited (16). Additionally, in colorectal cancer tumor tissue, the high expression of SERPINH1 was found to be significantly correlated with a high T stage, lymph node metastasis, venous infiltration, and a high TNM stage, indicating that SERPINH1 may be a new biomarker for colorectal cancer and is closely linked to its prognosis (15). However, to date, no studies had been conducted on the expression of SERPINH1 in LUAD and its potential prognostic relevance.
In the present study, we collected information on LUAD from the TCGA database to investigate the expression and survival prognosis of SERPINH1. Further, we analyzed the genes co-expressed with it, carried out an enrichment analysis of the GO and KEGG pathways, and further investigated the mechanism of this gene for the generation of LUAD. To better understand the mechanism of SERPINH1 in LUAD development, we then examined the role of SERPINH1 in the LUAD TMB, immunological microenvironment, immune infiltration, immune checkpoint inhibitors, and therapeutic sensitivity.
We found that the expression of SERPINH1 varied between LUAD tissues and normal tissues, and was highly expressed in tumor tissues. The Kaplan-Meier survival curves showed that the low-expression group had a greater chance of survival than the high-expression group. To further explore the relationship between SERPINH1 and OS, we performed univariate and multivariate Cox analyses on both the TCGA cohort and the NTU cohort. The combined analysis of the two groups showed that SERPINH1 can be employed as a standalone predictor of prognosis in LUAD. To further explore the biological function of SERPINH1, we performed co-expression and functional enrichment analyses of this gene. According to the KEGG enrichment analysis, SERPINH1 is mostly linked to metabolic processes, cell proliferation, and migration. According to the GO enrichment pathway analysis, the ECM is the main SERPINH1 pathway enriched in LUAD. The ECM is part of the TME. A study has shown that TME affects the growth of malignant tumors and their treatment (49). Without the ECM, cellular processes, such as growth, proliferation, migration, and differentiation, cannot be accomplished (50). SERPINH1 is mainly involved in the synthesis of type I collagen, which is a major component of the ECM (51,52) and plays a major role in tissue fibrosis (53). Fibroblast ECM proteins primarily create a barrier around tumor tissue, and when they are altered, tumors are highly susceptible to invasion and metastasis (54-56). Conversely, the KEGG enrichment analysis directly demonstrated that SERPINH1 is closely associated with cell proliferation and migration. We can infer from our findings that SERPINH1 is critically involved in the development of LUAD.
Research has shown a significant correlation between TMB and immunotherapy. The TMB quantifies the number of tumor mutations. The higher the number of tumor mutations, the more immunogenicity is exposed by the tumor, which can increase the chance of recognition by T cells, which in turn increases the immunotherapeutic effect (57). As a result, it has been demonstrated that the TMB can be employed as a tumor immunotherapy prediction marker (38). Thus, we further explored the correlation between SERPINH1 and TMB. The results showed that the TMB was positively correlated with SERPINH1 gene expression, such that patients with high TMB and high SERPINH1 expression had a better survival advantage, and patients with low TMB and high SERPINH1 expression had the worst survival advantage, which is consistent with the other results.
Further, by calculating the somatic mutation data, we found that TP53 had the highest mutation frequency (47%) in the high SERPINH1 expression group, followed by TTN (43%) and MUC16 (41%). Conversely, the driver gene with the highest rate of mutation in the group with low SERPINH1 expression was TP53 (40%). TP53 is an important regulator of the cell cycle, cell differentiation, and cell death (58). Studies have shown that TP53 mutations in LUAD are closely associated with tumor immunotherapy (59,60). Our study suggests that patients with TP53 mutations in the SERPINH1 high expression group may derive greater benefits when treated with immune checkpoint inhibitors. These findings suggest that differences in the distribution of SERPINH1-associated somatic mutation driver genes are significantly associated with anti-tumor immunity and that the complex regulatory mechanisms of their interactions could provide new directions for immunotherapy in LUAD.
To further explore the potential mechanisms and complex role of SERPINH1 in the tumor immune microenvironment (TIME), we used the ESTIMATE algorithm to score immune cells and stromal cells in the TME, and the results showed that the SERPINH1 high expression group had higher stromal scores and ESTIMATE scores. We then scored 22 common immune cells using the CIBERSORT algorithm and showed that immune cells, such as CD8+ T cells, follicular helper T cells, and monocytes, had a higher abundance of immune infiltration in the SERPINH1 low expression group. Additionally, the SERPINH1 high expression group exhibited a greater immunological score in terms of regulatory T cells, M0 macrophages, and activated mast cells than the SERPINH1 low expression group. We also used the ssGSEA algorithm to assess 29 common immune features. The results showed that immune features, such as antigen presentation, chemokines, and immune checkpoints, were more highly expressed in the SERPINH1 high expression group than the SERPINH1 low expression group. Thus, we hypothesize that immunotherapy may have greater benefits for patients with high expressions of SERPINH1. In conclusion, SERPINH1 plays a non-negligible role in TIME.
Over the past decade, immune checkpoint inhibitors have become the standard of care for the treatment of non-small cell lung cancer (61). Immunotherapy outperforms traditional chemotherapy in terms of response and tolerability, and OS time (62). Thus, we explored the correlation between SERPINH1 and immune checkpoints. First, we analyzed the expression levels of common immune checkpoint genes. The results showed that most immune checkpoint genes were more highly expressed in the SERPINH1 high expression group than the SERPINH1 low expression group. The subsequent correlation analysis confirmed that SERPINH1 was positively correlated with most of the immune checkpoints. All results above predict a better immunotherapeutic outcome in the SERPINH1 high expression group and are consistent with the immune signature results obtained from our ssGSEA. Finally, we explored the relationship between SERPINH1 and common clinical anti-tumor drugs. We found that the IC50 values were lower in the high-expression group than in the low-expression group, which means that chemotherapeutic agents are likely to produce better treatment benefits for patients with high expressions of SERPINH1. These findings imply that immunotherapy and chemotherapy are more effective for patients with high expressions of SERPINH1, and that SERPINH1 may be a useful marker for anticipating immunotherapy and chemotherapy outcomes in LUAD.
There is no conflict between the worse prognosis in higher SERPINH1-expression population and better immunotherapy and anti-tumor drug treatment in high SERPINH1-expression population. Because the effect of immunotherapy and anti-tumor drug treatment is affected by many factors. Factors affecting immunotherapy include TMB (63), DNA mismatch repair defects (64), microsatellite instability (65), tumor microenvironment (66,67), etc. The efficacy of antitumor drugs depends on the individual genetic differences (68), the sensitivity to chemotherapy, and the drug concentration in vivo (69). The effect of high SERPINH1 expression on immunotherapy and anti-tumor drug treatment is only one aspect of judging the prognosis.
Conclusions
In summary, we comprehensively assessed the prognosis of SERPINH1 and LUAD and their relationship to immunotherapy and chemotherapy and obtained good validation results from several perspectives. Thus, SERPINH1 is an ideal and stable predictor for the clinical treatment of patients with LUAD. However, this study still has its limitations, and studies involving prospective cohorts need to be conducted to demonstrate the reliability of SERPINH1. Additionally, the in vivo and in vitro experimental validation of SERPINH1 is needed to fully understand the unique role of SERPINH1 in the development of LUAD and its regulatory mechanisms.
Acknowledgments
Funding: This study was supported by grants from the Nantong Science and Technology Project (No. JC2021090) and the Scientific Research Fund of Anlotinib Resistant Gastric Cancer (No. HXKT20211015).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1518/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1518/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1518/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Affiliated Hospital of Nantong University (No. 2022-L094). Informed consent was obtained from the patients to use their human tissue for this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer 2019;10:3-7. [Crossref] [PubMed]
- Oliver AL. Lung Cancer: Epidemiology and Screening. Surg Clin North Am 2022;102:335-44. [Crossref] [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [Crossref] [PubMed]
- Lee TY, Huang KY, Chuang CH, et al. Incorporating deep learning and multi-omics autoencoding for analysis of lung adenocarcinoma prognostication. Comput Biol Chem 2020;87:107277. [Crossref] [PubMed]
- Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 2018;9:117. [Crossref] [PubMed]
- Sihoe ADL. Video-assisted thoracoscopic surgery as the gold standard for lung cancer surgery. Respirology 2020;25:49-60. [Crossref] [PubMed]
- Rossi A, Di Maio M. Platinum-based chemotherapy in advanced non-small-cell lung cancer: optimal number of treatment cycles. Expert Rev Anticancer Ther 2016;16:653-60. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Reck M, Remon J, Hellmann MD. First-Line Immunotherapy for Non-Small-Cell Lung Cancer. J Clin Oncol 2022;40:586-97. [Crossref] [PubMed]
- Duarte BDP, Bonatto D. The heat shock protein 47 as a potential biomarker and a therapeutic agent in cancer research. J Cancer Res Clin Oncol 2018;144:2319-28. [Crossref] [PubMed]
- Qi Y, Zhang Y, Peng Z, et al. SERPINH1 overexpression in clear cell renal cell carcinoma: association with poor clinical outcome and its potential as a novel prognostic marker. J Cell Mol Med 2018;22:1224-35. [Crossref] [PubMed]
- Li Y, Deng G, Qi Y, et al. Bioinformatic Profiling of Prognosis-Related Genes in Malignant Glioma Microenvironment. Med Sci Monit 2020;26:e924054. [Crossref] [PubMed]
- Yang B, Zhang M, Luo T. Identification of Potential Core Genes Associated With the Progression of Stomach Adenocarcinoma Using Bioinformatic Analysis. Front Genet 2020;11:517362. [Crossref] [PubMed]
- Mori K, Toiyama Y, Okugawa Y, et al. Preoperative heat shock protein 47 levels identify colorectal cancer patients with lymph node metastasis and poor prognosis. Oncol Lett 2020;20:333. [Crossref] [PubMed]
- Yamamoto N, Kinoshita T, Nohata N, et al. Tumor-suppressive microRNA-29a inhibits cancer cell migration and invasion via targeting HSP47 in cervical squamous cell carcinoma. Int J Oncol 2013;43:1855-63. [Crossref] [PubMed]
- Wang F, Xue Q, Xu D, et al. Identifying the hub gene in gastric cancer by bioinformatics analysis and in vitro experiments. Cell Cycle 2020;19:1326-37. [Crossref] [PubMed]
- Nissen NI, Karsdal M, Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J Exp Clin Cancer Res 2019;38:115. [Crossref] [PubMed]
- Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 2012;196:395-406. [Crossref] [PubMed]
- Song K, Yu Z, Zu X, et al. Collagen Remodeling along Cancer Progression Providing a Novel Opportunity for Cancer Diagnosis and Treatment. Int J Mol Sci 2022;23:10509. [Crossref] [PubMed]
- Provenzano PP, Inman DR, Eliceiri KW, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med 2008;6:11. [Crossref] [PubMed]
- Zhu J, Xiong G, Fu H, et al. Chaperone Hsp47 Drives Malignant Growth and Invasion by Modulating an ECM Gene Network. Cancer Res 2015;75:1580-91. [Crossref] [PubMed]
- Taguchi T, Razzaque MS. The collagen-specific molecular chaperone HSP47: is there a role in fibrosis? Trends Mol Med 2007;13:45-53. [Crossref] [PubMed]
- Saitoh M. Epithelial-Mesenchymal Transition by Synergy between Transforming Growth Factor-β and Growth Factors in Cancer Progression. Diagnostics (Basel) 2022;12:2127. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Tian S, Peng P, Li J, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY) 2020;12:3574-93. [Crossref] [PubMed]
- Zhong H, Wang Z, Wei X, et al. Prognostic and immunological role of SERPINH1 in pan-cancer. Front Genet 2022;13:900495. [Crossref] [PubMed]
- Neophytou CM, Panagi M, Stylianopoulos T, et al. The Role of Tumor Microenvironment in Cancer Metastasis: Molecular Mechanisms and Therapeutic Opportunities. Cancers (Basel) 2021;13:2053. [Crossref] [PubMed]
- Noman MZ, Hasmim M, Lequeux A, et al. Improving Cancer Immunotherapy by Targeting the Hypoxic Tumor Microenvironment: New Opportunities and Challenges. Cells 2019;8:1083. [Crossref] [PubMed]
- Gedaly R, Cornea V, Turcios L, et al. Anti-neoplastic sulfonamides alter the metabolic homeostasis and disrupt the suppressor activity of regulatory T cells. Sci Rep 2022;12:19112. [Crossref] [PubMed]
- Pernot S, Evrard S, Khatib AM. The Give-and-Take Interaction Between the Tumor Microenvironment and Immune Cells Regulating Tumor Progression and Repression. Front Immunol 2022;13:850856. [Crossref] [PubMed]
- Han S, Wang W, Wang S, et al. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics 2021;11:2892-916. [Crossref] [PubMed]
- Codrici E, Popescu ID, Tanase C, et al. Friends with Benefits: Chemokines, Glioblastoma-Associated Microglia/Macrophages, and Tumor Microenvironment. Int J Mol Sci 2022;23:2509. [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Kumar V, Donthireddy L, Marvel D, et al. Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell 2017;32:654-68.e5. [Crossref] [PubMed]
- Bos PD, Plitas G, Rudra D, et al. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med 2013;210:2435-66. [Crossref] [PubMed]
- Mantovani A, Sica A, Sozzani S, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004;25:677-86. [Crossref] [PubMed]
- Mills CD, Kincaid K, Alt JM, et al. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 2000;164:6166-73. [Crossref] [PubMed]
- Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet 2021;398:1002-14. [Crossref] [PubMed]
- Zhou F, Qiao M, Zhou C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell Mol Immunol 2021;18:279-93. [Crossref] [PubMed]
- Schabath MB, Cote ML. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol Biomarkers Prev 2019;28:1563-79. [Crossref] [PubMed]
- Nooreldeen R, Bach H. Current and Future Development in Lung Cancer Diagnosis. Int J Mol Sci 2021;22:8661. [Crossref] [PubMed]
- Mori K, Toiyama Y, Otake K, et al. Proteomics analysis of differential protein expression identifies heat shock protein 47 as a predictive marker for lymph node metastasis in patients with colorectal cancer. Int J Cancer 2017;140:1425-35. [Crossref] [PubMed]
- Henke E, Nandigama R, Ergün S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Front Mol Biosci 2019;6:160. [Crossref] [PubMed]
- Girigoswami K, Saini D, Girigoswami A. Extracellular Matrix Remodeling and Development of Cancer. Stem Cell Rev Rep 2021;17:739-47. [Crossref] [PubMed]
- McKee TJ, Perlman G, Morris M, et al. Extracellular matrix composition of connective tissues: a systematic review and meta-analysis. Sci Rep 2019;9:10542. [Crossref] [PubMed]
- Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci 2010;123:4195-200. [Crossref] [PubMed]
- Karamanos NK, Piperigkou Z, Passi A, et al. Extracellular matrix-based cancer targeting. Trends Mol Med 2021;27:1000-13. [Crossref] [PubMed]
- Provenzano PP, Eliceiri KW, Campbell JM, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med 2006;4:38. [Crossref] [PubMed]
- Conklin MW, Eickhoff JC, Riching KM, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol 2011;178:1221-32. [Crossref] [PubMed]
- Zunder SM, Gelderblom H, Tollenaar RA, et al. The significance of stromal collagen organization in cancer tissue: An in-depth discussion of literature. Crit Rev Oncol Hematol 2020;151:102907. [Crossref] [PubMed]
- Jardim DL, Goodman A, de Melo Gagliato D, et al. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021;39:154-73. [Crossref] [PubMed]
- Pelosi G. KEAP1 and TP53 (Co)mutation in Lung Adenocarcinoma: Another Bullet for Immunotherapy? J Thorac Oncol 2021;16:1979-83. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Li X, Xu Y, Gong F, et al. 1319P Interdependence of KRAS and TP53 mutations in predicting benefit from immune checkpoint inhibitor (ICI) in non-squamous NSCLC. Ann Oncol 2020;31.
- Brozos-Vázquez EM, Díaz-Peña R, García-González J, et al. Immunotherapy in nonsmall-cell lung cancer: current status and future prospects for liquid biopsy. Cancer Immunol Immunother 2021;70:1177-88. [Crossref] [PubMed]
- Kang J, Zhang C, Zhong WZ. Neoadjuvant immunotherapy for non-small cell lung cancer: State of the art. Cancer Commun (Lond) 2021;41:287-302. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Bever KM, Le DT. DNA repair defects and implications for immunotherapy. J Clin Invest 2018;128:4236-42. [Crossref] [PubMed]
- Zhao P, Li L, Jiang X, et al. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol 2019;12:54. [Crossref] [PubMed]
- Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol 2016;27:1482-92. [Crossref] [PubMed]
- Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res 2019;79:4557-66. [Crossref] [PubMed]
- Ippolito MR, Martis V, Martin S, et al. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev Cell 2021;56:2440-54.e6. [Crossref] [PubMed]
- An J, Peng C, Tang H, et al. New Advances in the Research of Resistance to Neoadjuvant Chemotherapy in Breast Cancer. Int J Mol Sci 2021;22:9644. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


