
A multicenter-retrospective cohort study of chromosome instability in lung cancer: clinical characteristics and prognosis of patients harboring chromosomal instability detected by metagenomic next-generation sequencing
Highlight box
Key findings
• The median OS differed significantly between the 5p15dup+ group and the combined group in patients with lung cancer.
What is known and what is new?
• CIN were reported to correlate with cancer stage when diagnosed and with disease progression and poor prognosis.
• A real-world clinical characteristics and prognosis of patients in lung cancer harboring CIN, using mNGS to detect.
What is the implication, and what should change now?
• Various forms of CIN detected by mNGS may predict prognosis of patients with lung cancer differentially.
• CIN with duplication or deletion deserves further study to guide clinical treatment.
Introduction
Lung cancer is the leading cause of cancer-related deaths (1). Traditionally, lung cancer diagnosis has been determined by pathology, polymerase chain reaction (PCR), or immunohistochemistry (IHC) methods. More recently, this has also included novel next-generation sequencing (NGS) detection. However, recurrence after surgical resection, disease progression, and resistance to anti-tumor therapy (including targeted therapy and immunotherapy) remain major unsolved problems. In the search for novel predictors for early diagnosis, rapid technological advances in liquid biopsy analyses, such as circulating tumor DNA (ctDNA) and cell-free DNA (cfDNA), have come to the forefront as auxiliary cancer diagnostic methods (2-4).
Extensive efforts have focused on the genomic features of primary or metastatic lung cancer. One form of genomic instability, chromosomal instability (CIN), a hallmark of human cancer, refers to genomic alterations that contain chromosomal number alterations or structural aberrations, ranging from single nucleotide mutations to whole chromosome changes, such as aneuploidy (5,6). CIN can induce tumor cell survival and metastases by upregulation of inflammatory pathways, resulting in complex consequences in cellular-level mechanisms (7,8). Previous research has confirmed the links between CIN and disease stage, metastasis, poor prognosis, and therapeutic resistance (9-11). However, refers to lung cancer, real-world clinical characteristics and prognosis in patients with CIN-positive and CIN-negative remains unclear. It is meaningful to seek the relationship between CIN forms and clinical outcomes for disease assessment.
Novel sequencing and analytical methods greatly facilitate the identification and presentation of chromosomal copy number variants (CNVs) and provide more possibilities to explore the dynamic process of CIN. Chromosomal deletions or duplications can be explored by metagenomic NGS (mNGS), which has been widely used to distinguish various pathogens like fungi, viruses, and mycoplasma (12). CIN detected by mNGS, which applies the human reads to map the reference human database in lung biopsy tissue, showed that mNGS had a clinical sensitivity of 83.7%, a specificity of 97.6%, and a 92.9% accuracy compared with the results of pathological examinations (13). Power of CIN collected from BALF predict cancer remains unknown. A study correlating breast cancer with CIN showed that different molecular types harbored different CNVs in their chromosomes (14).
Therefore, this study aimed to demonstrate clinical characteristics and prognosis of patients in lung cancer harboring CIN analyze the sensitivity and specificity of mNGS detecting CIN in diagnosing cancer. Furthermore, we designed this study to explore the molecular karyotype categorization and clinical characteristics of lung cancer patients harboring positive CIN and whether positive CIN detected by mNGS predicted therapy effects in patients with lung cancer. We present the following article in accordance with the STARD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1732/rc).
Methods
Patient enrollment and clinical assessment
This was a multicenter, retrospective cohort study in which patients were enrolled from ten general hospitals in Fujian, China, between January 2021 and January 2022 upon preliminary diagnosis of a suspected pulmonary infection or lung cancer. Hospitals contained the 900th Hospital of the Joint Logistic Support Force, Fujian Provincial Hospital, the Second Affiliated Hospital of Fujian Medical University, the First Affiliated Hospital of Fujian Medical University, Union Hospital Affiliated to Fujian Medical University, the Affiliated People’s Hospital of Fujian University of Traditional Chinese Medicine, Fuzhou Pulmonary Hospital, Mindong Hospital of Ningde City, Affiliated Hospital of Putian University and Quanzhou First Hospital Affiliated to Fujian Medical University. Whole patients were between 18 and 83 years of age. From bronchoalveolar lavage fluid (BALF), tissue, blood, pleural, marrow, and sputum samples, clinical cases detected by mNGS were consecutively obtained from 668 patients recruited into the study. Among the 668 cases, 81 were CIN-positive, and 587 were CIN-negative. There were 22 cases with missing clinical information, and six remained diagnostically undetermined until our endpoint. In 81 CIN-positive cases, 53 cases were cancers determined by clinical pathology including 27 cases of lung cancer. A diagnosis of lung cancer determined by pathology was established in 46 cases in total, 27 from the CIN-positive group and 19 from the CIN-negative group. Four patients were excluded from the survival analysis due to loss or absence of treatment information. The resulting 42 cases and their associated demographic and clinical data were included in the final analysis. Baseline clinical data contained age, sex, smoking status, history, pathologic type, stage, lung metastases (Figure 1). The patients were followed-up from registered to September 2022, no matter reach outcome event. Responses were measured according to the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST 1.1).
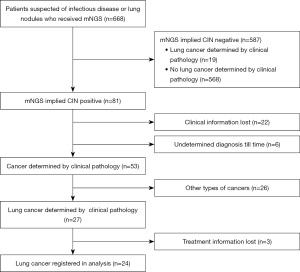
Integrated diagnosis included: (I) pathology confirmed or excluded (Lung cancer were staged according to American Joint Committee on Cancer, 8th edition); (II) image evaluation implied that lesion lessened; (III) relief of clinical symptoms after anti-infectious treatment, such as cough, expectoration, fever, and dyspnea; (IV) pathogens detected by conventional detection or mNGS, such as bacteria, fungi, tuberculosis, and virus. Following the listed integrated information, we distinguished patients with and without lung cancer. The clinical information and reference standard results were available to the performers/readers of the index test. The clinical information and index test results were available to the assessors of the reference standard.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the 900th Hospital of the Joint Logistic Support Force (No. 2022-028). Other hospitals were informed and agreed with this study. Individual consent for this retrospective analysis was waived.
BALF collection and DNA extraction
Samples from BALF, tissue, and pleura were collected from patients who had received bronchoscopic or computed tomography (CT)-guided transthoracic core needle biopsy (TNB). Samples were stored at 25 ℃ and sent to the library immediately. cfDNA from specific patients was trimmed and matched with human reads by whole exome sequencing (WES). The cancer pipeline was identified, and the human reads were counted using the sliding window technique of human genomic sequencing (15-17).
Gene-level copy number analyses by mNGS
Microbiology and malignancy screens were detected after cfDNA was extracted. The reserved human reads aligned with the human genomic sequencing and mapped reads were accessed for deeper analysis. The human genomic sequencing was dissected into continuous windows with a fixed length to detect the read depth of each window. Then, through normalization and smoothing, chromosome position and CIN, including depletion and duplication, were obtained. Finally, the copy number of segments on aberrant chromosomes was calculated, and CNVs were validated with the setting thresholds (18).
Samples that were CIN-negative as detected by mNGS were repeatedly analyzed to ensure that no CIN had been detected and the results were negative. The clinical lavage sites were matched with the tumor positions.
NGS/PCR
Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue or blood. Samples were analyzed at the central laboratory for NGS detection (Amoy Diagnostics, Xiamen, China). Extracted DNA underwent fragmentation, terminal modification, ligation of adapters, library amplification, hybridization capture, primer synthesis, and sequencing to acquire mutation status (19). Real-time quantitative PCR (RT-qPCR) is a homogeneous method to acquire amplification information and analysis after the fluorescence of DNA dyes and probes are tested by each PCR cycle. After accumulation of the cycle reaches a specific number, fluorescence exceeds that background. The point is quantified as the second derivative maximum (crossing point) and influences the starting copy of the PCR reaction (20).
Statistical analyses
In CIN-positive group and combined group, as well as comparing Chr5p15 dup group to combined group, the Student’s t-test and the chi-square test were used to calculate differences in continuous and categorical data, respectively. With the significance level of 0.05, we use PASS 15 Power Analysis and Sample Size Software (NCSS, LLC. Kaysville, UT, USA) to finish the power analyses with a two-sided test in a logistic regression. The receiver operating characteristic (ROC) curve was used to identify the cut-off value and estimate measures of diagnostic accuracy. Survival curves were analyzed by the Kaplan-Meier method and compared using the Wilcoxon test. Statistical significance was set at P<0.05. All statistical analyses were performed with SPSS Statistics 25.0 software (IBM Corporation, Armonk, NY, USA).
Results
Sensitivity and specificity of cancer diagnosis using mNGS from BALF
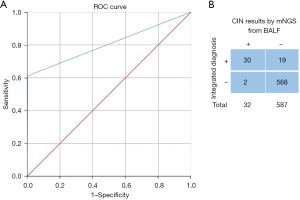
To assess the probability of CIN predicting cancer, we analyzed BALF samples collected by bronchoscopy in 619 cases based on conventional pathology detection methods like PCR, IHC, and cytology. The mNGS and conventional detection were processed simultaneously. Of the 619 patients, 30 were confirmed to have malignancy on histology, and two cases remained undetermined lung nodules until our endpoint and were classified as false positive reactions. By integrated diagnosis, 587 patients were excluded with benign lesions (Figure S1). The cut-off values were established based on the ROC curve [area under the curve (AUC) =0.804; 95% confidence interval (CI), 0.719–0.89; Figure 2A]. Compared with the results of the pathological examination, the mNGS had a clinical sensitivity of 61.22%, a specificity of 99.65%, and 83.17% accuracy (Figure 2B). There were no adverse events from performing the index test or the reference standard.
Patient characteristics
In the 42 patients with pathologist-confirmed lung cancer diagnoses, mNGS detected 24 patients as CIN-positive and 18 as CIN-negative. The clinical and pathological characteristics of the patients are detailed in Table 1. In these individuals, the mean age at lung cancer diagnosis was 64.17 years in the CIN-positive group (range, 33–83 years), compared to 60.22 years in the CIN-negative group (range, 30–82 years; P=0.466). In the CIN-positive group, 18 out of 24 patients (75.0%) were unresectable differentiated stage III or IV at primary diagnosis, compared to 11 out of 18 (61.11%) in the CIN-negative group. Of the tumors harboring CIN, there were 19 adenocarcinomas, three lung squamous cell carcinomas (LSCC), and two small-cell lung cancers (SCLCs). One pathologist reviewed the histological diagnoses.
Table 1
| Characteristics | CIN-positive (n=24) | CIN-negative (n=18) | P |
|---|---|---|---|
| Age (years) | 64.17±12.83 | 60.22±12.54 | 0.325 |
| Sex, n (%) | 0.582 | ||
| Male | 14 (58.33) | 12 (66.67) | |
| Female | 10 (41.67) | 6 (33.33) | |
| Smoking, n (%) | 11 (45.83) | 10 (55.56) | 0.533 |
| History, n (%) | 0.072 | ||
| Cancer | 2 (8.33) | 0 | |
| Lung disease | 2 (8.33) | 6 (33.33) | |
| Except disease up | 20 (83.34) | 12 (66.67) | |
| Pathology, n (%) | 0.366 | ||
| Adenocarcinoma | 19 (79.17) | 11 (61.11) | |
| LSCC | 3 (12.5) | 5 (27.77) | |
| SCLC | 2 (8.33) | 1 (5.56) | |
| Lymphoepithelioid, n (%) | 0 | 1 (5.56) | |
| Stage, n (%) | 0.142 | ||
| I | 3 (12.5) | 6 (33.33) | |
| II | 3 (12.5) | 1 (5.56) | |
| III | 2 (8.33) | 4 (22.22) | |
| IV | 16 (66.67) | 7 (38.89) | |
| Lung metastasis, n (%) | 10 (41.67) | 7 (38.89) | 0.856 |
The ages were accounted with mean ± standard deviation. The age difference was calculated by independent t-test. Statistical data of sex, smoking, history, pathology, stage, and lung metastasis were calculated by the chi-square test. CIN-positive: patients with CIN detected by mNGS; CIN-negative: patients without CIN determined by mNGS; history: disease history; lung disease: pneumonia, tuberculosis, interstitial pneumonia, respiratory failure, and chronic obstructive pulmonary disease. CIN, chromosomal instability; LSCC, lung squamous cell carcinoma; SCLC, small-cell lung cancer; mNGS, metagenomic next-generation sequencing.
Characteristics of CIN distribution in chromosomes
Twenty-seven cases were identified as CIN-positive, but two were excluded due to a lack of information integrity. The 25 lung cancer cases with CIN-positive status, including the different clinical stages ranging from I to IV, are listed in Figure 3A. In these 25 cases, there were 523 chromosomal CNV changes, with forms including duplication (dup), deletion (del), mosaic (mos), and whole chromosome amplification or loss (Figure 3B). A total of 243 duplication variants and 192 deletion variants occurred in all chromosomes. Duplications occurred in most chromosomes except for Chr9 and Chr13, in which CNV tended to delete. For patients with chromosomal duplications, the most frequent CNVs were 1q21 (8/243), 5p15 (12/243), 7p22 (9/243), and 8q24 (8/243). For patients with chromosomal deletions, the most frequent CNVs were 8p23 (10/192), 17p13 (10/192), and 19p13 (8/192). There was no difference among the frequency ratios (Kruskal-Wallis test, P=0.460). Information on clinical databases, including overall survival (OS) status, was available for 17 patients with unresectable cancer stage III/IV (Table 2). The median OS in patients with Chr5p15 duplication was 32.4 months (95% CI, 10.35–54.45 months). There was a significant difference in the median OS between the 5p15dup+ group and the combined group (32.4 vs. 8.63 months, P=0.049; Figure 3C).
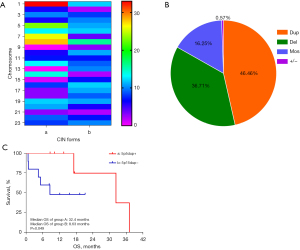
Table 2
| Characteristics | 5p15dup+ (n=7) | 5p15dup− (n=10) | P |
|---|---|---|---|
| Age (years) | 59.57±13.25 | 66.50±13.18 | 0.304 |
| Male ratio, n (%) | 4 (57.1) | 7 (70.0) | 0.664 |
| Pathology, n (%) | 0.394 | ||
| Adenocarcinoma | 6 (85.7) | 6 (60.0) | |
| LSCC | 1(14.3) | 2 (20.0) | |
| SCLC | 0 | 2 (20.0) | |
| Gene mutation, n (%) | 3 (42.9) | 4 (40.0) | 1.000 |
| Stage, n (%) | 0.485 | ||
| III | 0 | 2 (20.0) | |
| IV | 7 (100.0) | 8 (80.0) |
Duplication and deletion occurred with various cumulative frequencies in all chromosomes in 25 patients. Seventeen patients with intact follow-up information were recruited to analyze the difference in prognosis in the 5p15dup+ and 5p15dup− groups. The ages were accounted with mean ± standard deviation. The age difference was calculated with an independent t-test. Statistical data of sex, pathology, gene mutation, and stage were calculated by the chi-square or Fisher’s exact test. LSCC, lung squamous cell carcinoma; SCLC, small-cell lung cancer.
Prognostic analyses of unresected lung cancer with CIN-positive status
Of the 24 CIN-positive patients, samples from BALF and tissue comprised 62.5% and 25%, respectively. Pleural and blood samples comprised 4.17% and 8.33%, respectively (Figure 4A). Eighteen of these 24 patients were unresected. There were 11 patients with lung cancer in the CIN-negative group. Analysis was conducted of the total number of 29 patients with unresected lung cancer. In the CIN-positive group, the median progression-free survival (mPFS) of seven patients with adenocarcinoma receiving targeted therapy was 8.0 months; in contrast, the mPFS was 11.57 months in patients in the CIN-negative group (n=5). The mPFS was 3.8 months in patients with adenocarcinoma who received chemotherapy combined with immunotherapy and was 7.3 months and 2.2 months in patients with SCC and SCLC who received chemotherapy combined with immunotherapy, respectively. The detailed treatment responses are listed in Tables 3,4. The median OS of 18 cases in the CIN-positive group was 32.4 months (95% CI, 14.2–50.6 months), and the median OS of 11 cases in the CIN-negative group was 35.63 months (95% CI, 21.64–49.62 months; Wilcoxon, P=0.227; Figure 4B). In multiple adjustment analysis, CIN status, gender, age, smoking status, history, stage and lung metastases failed to predict the outcomes of patients.
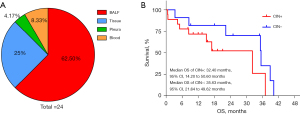
Table 3
| Pathology type | Therapy | Cases | mPFS (months) | PD | SD | PR |
|---|---|---|---|---|---|---|
| Adenocarcinoma (n=13) | Targeted | 7 | 8.00 | 1 | 2 | 4 |
| Chemo + IO | 2 | 3.80 | 0 | 2 | 0 | |
| None | 4 | – | – | – | – | |
| LSCC (n=3) | Chemo + IO | 2 | 7.30 | 0 | 2 | 0 |
| None | 1 | – | – | – | – | |
| SCLC (n=2) | Chemo + IO | 2 | 2.20 | 0 | 0 | 2 |
CIN, chromosomal instability; mPFS, median progression-free survival; PD, progressive disease; SD, stable disease; PR, partial response; targeted, targeted therapy; chemo, chemotherapy; IO, immunotherapy; none, did not accept anti-tumor therapy; LSCC, lung squamous cell carcinoma; SCLC, small-cell lung cancer.
Table 4
| Pathology type | Therapy | Cases | mPFS (months) | PD | SD | PR |
|---|---|---|---|---|---|---|
| Adenocarcinoma (n=5) | Targeted | 3 | 11.57 | – | 1 | 2 |
| Chemo + IO | 1 | – | – | 1 | – | |
| Chemo | 1 | 19.17 | – | – | 1 | |
| LSCC (n=4) | Chemo + IO | 3 | 8.23 | 1 | – | 2 |
| Chemo | 1 | 10.63 | – | 1 | – | |
| SCLC (n=1) | Chemo + IO | 1 | 3.80 | – | 1 | – |
| Lymphoepithelioid (n=1) | Chemo + IO | 1 | 14.87 | – | – | 1 |
CIN, chromosomal instability; mPFS, median progression-free survival; PD, progressive disease; SD, stable disease; PR, partial response; targeted, targeted therapy; chemo, chemotherapy; IO, immunotherapy; none, did not accept anti-tumor therapy; LSCC, lung squamous cell carcinoma; SCLC, small-cell lung cancer.
Discussion
CIN is a hallmark of human cancer. In our study, patients with lung cancer harboring CIN from various sample origins were found to have poor prognosis and more metastasis than CIN-negative patients, but there was no statistical difference between the groups. Of the lung cancer patients with CIN-positive status, 66.67% were stage IV compared to 38.89% in the CIN-negative status group. Multiple metastases and OS differed from that in CIN-negative lung cancer, but there was no statistical difference, as this study revealed. Targeted therapies, immunotherapies, and chemotherapies combined with immunotherapies have significantly improved patient outcomes (21,22). However, despite aggressive interventions, many patients experience disease progression or acquire resistance to treatment. The immediate consequence of CIN is aneuploidy, which is the hallmark of aggressive malignancies. A higher aneuploidy burden and CIN were reported to correlate with cancer stage when diagnosed and with disease progression and poor prognosis in prostate cancer, breast cancer, and glioma (23). For example, duplication on Chr20q was reported as a predictor of shorter OS in patients with endometrial cancer (24). Given the limited sample numbers, this study listed the CIN conditions detected by mNGS and suggested that the duplication of Chr5p15 may be associated with a longer OS in lung cancer. However, due to the small sample numbers, further replication of this finding is urgently required.
Copy number aberrations belonging to cancer include deletions and duplication at small segments, indels/single-nucleotide variations, gain or loss of chromosome arms, or whole chromosome and whole-genome doubling. Individual chromosomal aberrations have been correlated with mutational status in breast cancers (25). As reported, we displayed the whole expression of CNV in a registered cohort. In our study, the status of driver or typical genes in the CIN-positive group was 9/24 (37.5%) compared to 5/18 (27.78%) in the CIN-negative group. In gastric cancer, the correlation between driver genes and CNVs showed that CNVs provide rich information that may reflect disease-related signaling patterns and clinicopathological features, so more research on the association between CNVs and driver genes in lung cancer may improve the curative effects of targeted therapy (26).
In a previous study, CIN detected by mNGS by applying the human reads to map the reference human database in lung biopsy tissue showed that mNGS had a clinical sensitivity of 83.7%, a specificity of 97.6%, and 92.9% accuracy compared with pathological examination results (13). In contrast, our study showed that mNGS had a clinical sensitivity of 61.22%, a specificity of 99.65%, and 83.17% accuracy compared with pathological examination results. One possible reason for this difference in results may be due to collecting the BALF samples by bronchoscopy in our study. In CIN-negative group, six of 18 (33.33%) patients with lung cancer were at stage I, compared with 3/24 (12.5%) in CIN-positive group. The negative detection result implied that acquiring CIN from BALF was more challenging in early-stage lung cancer. Conversely, more CIN information from BALF was acquired in advanced-stage lung cancer. The other reason may be that samples gathered from lung tissues obtain better sensitivity than BALF. However, since the specificity was significant in our study, CIN detected from BALF samples still provides clinicians with a novel way to distinguish benign vs. malignant lung nodules. Our future research efforts will include a larger sample size to investigate the differences between BALF and lung tissue samples in detecting CIN in early-stage lung cancer. Additionally, the loss of some clinical information may have affected the statistical accuracy, and the limitation of sample origins indicates the need for further research.
We acquired a database of CIN detected from BALF, pleura, blood, lung tissue, and sputum samples to determine the presence of cancer. The results suggest that mNGS can provide clinicians with a rapid and accurate method of diagnosing lung cancer. But the significance of different CIN patterns requires further research.
Conclusions
In this study, we demonstrated the clinical characteristics and prognostic differences between CIN-positive and CIN-negative lung cancer groups. By extracting CIN data from BALF, mNGS may be useful in predicting the possibility of lung cancer. CIN with duplication or deletion deserves further study to guide clinical treatment.
Acknowledgments
Thanks to all hospitals for the support with information of enrolled cases (Fujian Provincial Hospital, the Second Affiliated Hospital of Fujian Medical University, the First Affiliated Hospital of Fujian Medical University, Union Hospital Affiliated to Fujian Medical University, the Affiliated People’s Hospital of Fujian University of Traditional Chinese Medicine, Fuzhou Pulmonary Hospital, Mindong Hospital of Ningde City, Affiliated Hospital of Putian University and Quanzhou First Hospital Affiliated to Fujian Medical University included).
Funding: This work was supported by the Fujian University of Traditional Chinese Medicine: Scientific Research Program of University Management (Grant No. XB2022142) and the External Cooperation of Science and Technology Program of Fujian Province (No. 202210034).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1732/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1732/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1732/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the 900th Hospital of the Joint Logistic Support Force (No. 2022-028). Other hospitals were informed and agreed with this study. Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Mathios D, Johansen JS, Cristiano S, et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun 2021;12:5060. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Xia L, Mei J, Kang R, et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin Cancer Res 2022;28:3308-17. [Crossref] [PubMed]
- Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol 2018;15:139-50. [Crossref] [PubMed]
- Chan SH, Ngeow J. Germline mutation contribution to chromosomal instability. Endocr Relat Cancer 2017;24:T33-46. [Crossref] [PubMed]
- Hong C, Schubert M, Tijhuis AE, et al. cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 2022;607:366-73. [Crossref] [PubMed]
- Bakhoum SF, Cantley LC. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018;174:1347-60. [Crossref] [PubMed]
- Tijhuis AE, Johnson SC, McClelland SE. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol Cytogenet 2019;12:17. [Crossref] [PubMed]
- Bakhoum SF, Ngo B, Laughney AM, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467-72. [Crossref] [PubMed]
- Bach DH, Zhang W, Sood AK. Chromosomal Instability in Tumor Initiation and Development. Cancer Res 2019;79:3995-4002. [Crossref] [PubMed]
- Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med 2021;27:115-24. [Crossref] [PubMed]
- Guo Y, Li H, Chen H, et al. Metagenomic next-generation sequencing to identify pathogens and cancer in lung biopsy tissue. EBioMedicine 2021;73:103639. [Crossref] [PubMed]
- Mo H, Wang X, Ma F, et al. Genome-wide chromosomal instability by cell-free DNA sequencing predicts survival in patients with metastatic breast cancer. Breast 2020;53:111-8. [Crossref] [PubMed]
- Fan HC, Blumenfeld YJ, Chitkara U, et al. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A 2008;105:16266-71. [Crossref] [PubMed]
- Talevich E, Shain AH, Botton T, et al. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12:e1004873. [Crossref] [PubMed]
- Gu W, Talevich E, Hsu E, et al. Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids. Genome Med 2021;13:98. [Crossref] [PubMed]
- Su J, Han X, Xu X, et al. Simultaneous Detection of Pathogens and Tumors in Patients With Suspected Infections by Next-Generation Sequencing. Front Cell Infect Microbiol 2022;12:892087. [Crossref] [PubMed]
- Chen Y, Xu J, Zhang L, et al. A multicenter-retrospective study of non-small-cell lung carcinoma harboring uncommon epidermal growth factor receptor (EGFR) mutations: different subtypes of EGFR exon 19 deletion-insertions exhibit the clinical characteristics and prognosis of non-small cell lung carcinoma. Transl Lung Cancer Res 2022;11:238-49. [Crossref] [PubMed]
- Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond) 2005;109:365-79. [Crossref] [PubMed]
- Baudino TA. Targeted Cancer Therapy: The Next Generation of Cancer Treatment. Curr Drug Discov Technol 2015;12:3-20. [Crossref] [PubMed]
- Lukow DA, Sheltzer JM. Chromosomal instability and aneuploidy as causes of cancer drug resistance. Trends Cancer 2022;8:43-53. [Crossref] [PubMed]
- Hieronymus H, Murali R, Tin A, et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018;7:e37294. [Crossref] [PubMed]
- Shukla A, Nguyen THM, Moka SB, et al. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat Commun 2020;11:449. [Crossref] [PubMed]
- Pernas S, Barroso-Sousa R, Tolaney SM. Optimal treatment of early stage HER2-positive breast cancer. Cancer 2018;124:4455-66. [Crossref] [PubMed]
- Liang L, Fang JY, Xu J. Gastric cancer and gene copy number variation: emerging cancer drivers for targeted therapy. Oncogene 2016;35:1475-82. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


