
High expression of FGFR3 predicts a better prognosis for patients with non-small cell lung cancer in a Chinese population
Highlight box
Key findings
• Our study showed that FGFR3 was highly expressed in NSCLC tissues, and the frequency rate for the FGFR3 mutation at T450 M in NSCLC tissues was low.
What is known and what is new?
• Several studies have detected alterations of FGFR3 in NSCLC; however, the frequency of FGFR3 status was discordance. In one study, FGFR3 fusions were detected in 9 of 312 lung SCCs in Chinese patients, and the rate of FGFR3 fusions was even lower in lung ACs (0.6%, 6/1,016).
• Thus, this study sought to examine the alterations of FGFR3 using immunohistochemistry and Sanger sequencing in the NSCLC tissues of 116 patients with surgically resectable NSCLC at the I-IIIA stage. The prognostic role of FGFR3 in NSCLC patients was also explored.
What is the implication, and what should change now?
• FGFR3 may serve as an independent prognostic factor for the OS of NSCLC patients.
Introduction
Lung cancer is a leading cause of cancer-related deaths worldwide (1). Non-small cell lung cancer (NSCLC) accounts for >75% of all cases of lung cancer and can be further classified into 3 subtypes; that is, adenocarcinoma (AC), squamous cell carcinoma (SCC), and large cell carcinoma (2-5). A number of studies have successfully applied molecular-targeted therapeutic agents and demonstrated their superiority to conventional chemotherapy in treating NSCLC (6-8). For example, the echinoderm microtubule-associated protein like4-anaplasticlymphoma kinase and epidermal growth factor receptor (EGFR) are important treatment modalities for NSCLC (9-12). Despite advances in the multimodality treatment of NSCLC, the prognosis of patients still remains unfavorable with a 5-year overall survival (OS) rate of <15% in China (13,14). To improve the prognosis of NSCLC patients and establish effective treatment modalities, the tumor biology mechanism needs to be deciphered.
Fibroblast growth factor receptors (FGFRs) are 4 highly conserved transmembrane receptor tyrosine kinase (RTK) families, and include FGFR1, FGFR2, FGFR3, and FGFR4 (15). FGFR3 spans 16.5 kb with 18 introns and 19 exons and is situated on chromosome 4p16.3. FGFR3 is a member of the FGFR family and has been found to regulate cell growth, cell differentiation, and migration (16,17). FGFR3 is involved in a variety of biological processes including cell proliferation, differentiation, migration, angiogenesis, and apoptosis. Cellular functions of FGFR3 are activated by ligand-induced dimerization which in turn leads to transphosphorylation of key tyrosine residues in the kinase domain of the receptor Phosphorylated tyrosines act as docking sites for the recruitment of signaling molecules and activation of downstream pathways. Four main signal transduction pathways have been implicated in mediating FGFR3 functions including the MAP kinase, STAT1, PI3K-AKT, and PLCγ (18,19). FGFR3 has been shown to be activated by the mutation or fusion of its own gene in several types of cancers, such as urinary bladder cancer, cervix carcinomas, glioblastoma, rhabdomyosarcoma, and NSCLC (20).
Several studies have detected alterations of FGFR3 in NSCLC (21-23); however, the frequency of FGFR3 status was discordance. In one study, FGFR3 fusions were detected in 9 of 312 lung SCCs in Chinese patients, and the rate of FGFR3 fusions was even lower in lung ACs (0.6%, 6/1,016) (23). In addition to FGFR3 fusions, the mutation alteration is more common in the tumor area, particularly in bladder tumors. A previous study reported that the FGFR3 mutation rate is rare in NSCLC, and is only found in lung SCC (21). To date, no data are available on the distribution rates for FGFR3 expression and mutation, its relation to clinicopathological characteristics, and clinical outcomes in NSCLC. Thus, this study sought to examine the alterations of FGFR3 using immunohistochemistry (IHC) and Sanger sequencing in the NSCLC tissues of 116 patients with surgically resectable NSCLC at the I-IIIA stage. The prognostic role of FGFR3 in NSCLC patients was also explored. The findings of the present study may provide novel insights into the role of FGFR3 in the pathophysiology of NSCLC. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1523/rc).
Methods
Patients
This study examined 116 NSCLC surgical tumor tissue samples from patients who had undergone primary and curative resection at the Affiliated Cancer Hospital and Institute of Guangzhou Medical University. The samples were collected between June 2010 and June 2014. None of the patients had previously undergone preoperative chemotherapy or radiotherapy. Patients with surgically resectable NSCLC (stage I–IIIA) were included in this study. The clinicopathological data of the patients are summarized in Table 1. Tumor, node and metastasis (TNM) staging was evaluated based on American Joint Committee on Cancer (AJCC)/International Union against Cancer TNM classification system (24). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Guangzhou Medical University (No. 50, 2019), and informed consent was obtained from each patient.
Table 1
| Variable | Case, N=86 (%) | FGFR3 expression | P value | |
|---|---|---|---|---|
| Low, N=26 (%) | High, N=26 (%) | |||
| Age (years) | 0.595 | |||
| ≥60 | 50 (58.8) | 36 (60.0) | 14 (53.8) | |
| <60 | 36 (41.2) | 24 (40.0) | 12 (46.2) | |
| Gender | 0.002 | |||
| Male | 57 (66.3) | 46 (76.7) | 11 (42.3) | |
| Female | 29 (33.7) | 14 (23.3) | 15 (57.7) | |
| Smoking | <0.001 | |||
| Ever-smoker | 42 (48.8) | 38 (63.3) | 4 (15.4) | |
| Never-smoker | 44 (51.2) | 22 (36.7) | 22 (84.6) | |
| Histology type | <0.001 | |||
| AC | 39 (44.2) | 17 (28.3) | 22 (84.6) | |
| SCC | 47 (55.8) | 43 (71.7) | 4 (15.4) | |
| T stage | 0.017 | |||
| ≤5 cm | 65 (75.6) | 41 (68.3) | 24 (92.3) | |
| >5 cm | 21 (24.4) | 19 (31.7) | 2 (17.7) | |
| N stage | 0.082 | |||
| 0 | 44 (51.2) | 27 (45.0) | 17 (65.4) | |
| 1, 2, 3 | 42 (48.8) | 33 (55.0) | 9 (34.6) | |
| Clinical stage | 0.934 | |||
| I, II | 59 (68.6) | 41 (66.7) | 18 (69.2) | |
| III | 27 (31.4) | 19 (33.3) | 8 (30.8) | |
| EGFR mutation | <0.001 | |||
| Yes | 39 (45.3) | 17 (28.3) | 22 (84.6) | |
| No | 47 (54.7) | 43 (71.7) | 4 (15.4) | |
NSCLC, non-small cell lung cancer; AC, adenocarcinoma; EGFR, epidermal growth factor receptor; FGFR3, fibroblast growth factor receptor 3; SCC, squamous cell carcinoma.
Tissue specimens
Formalin-fixed paraffin-embedded (FFPE) tissue blocks and the corresponding hematoxylin-and-eosin (H&E)-stained slides were overlaid for tissue microarray (TMA) sampling. The slides were assessed by a senior pathologist, who labelled the representative tumor areas. The TMA block (116 lung cancers tissues) was generated by punching representative tumor areas with cylinders (1 mm in diameter).
IHC analysis of NSCLC samples
The TMA slides were deparaffinized and rehydrated, and then incubated for 20 min with 3% hydrogen peroxide to quench endogenous peroxidase activity. Antigen retrieval was performed by 3-min pressure cooking in ethylenediaminetetraacetic acid buffer (pH 8.0). The slides were blocked with 10% normal goat serum for 30 min at room temperature. The slides were then incubated with rabbit FGFR3 monoclonal antibody (1:100, ab137084; Abcam, Cambridge, UK) for 2 h at room temperature, and then incubated with a secondary antibody for 1 h at room temperature. The staining was visualized with 3, 3'-diaminobenzidine. Normal brain and testicular tissues were used as positive control (see Figure S1), while negative control slides were generated by replacing the FGFR3 monoclonal antibody with normal immunoglobulin.
IHC staining evaluation
All the specimens were examined by 2 independent observers blinded to the clinicopathologic data. If inconsistencies arose, the cases were discussed until a consensus was reached. FGFR3-positive staining cancer cells were assessed at 100× magnification. Membranous and/or cytoplasmic staining was considered reactive. Intensity was scored as 3 for strong staining, 2 for moderate staining, 1 for weak staining, and 0 for negative staining. FGFR3 overexpression was defined by a score of 2 or 3. A higher score was selected for analysis if 2 discordant cores were generated for the same patient.
Evaluation of the FGFR3 mutation
A manual micro-dissection procedure on the tissue block was performed using H&E slides as templates. Based on the histological analysis, the dissected samples had >70% tumor cells. A DNAeasy® tissue kit was used for the deoxyribonucleic acid (DNA) extraction. The direct-sequencing method was used to analyze the FGFR3 mutation by choosing exons 7, 10, and 15 according to previous studies (25) and the primers used for the exon amplification have also been described previously (25). Polymerase chain reaction (PCR) products were fractionated by electrophoresis and visualized with ethidium bromide. The bigDye terminator V1.1 cycle sequencing kit was used to perform the sequencing analysis of the purified PCR products. ChromasPro software 1.34 was used for the mutational analysis.
Follow-up study
The follow-up period was defined as the date of diagnosis until any event (e.g., relapse, death, or last known follow-up). OS was defined as the date of diagnosis to tumour-related death. Disease-free survival (DFS) was defined as the date of diagnosis to tumor local recurrence or metastasis. The average follow-up period for the NSCLC patients was 36.46 months (median: 33.44 months; range, 4–45 months).
Statistical analysis
All the data were analyzed using SPSS software 16.0 (SPSS Inc., Amond, USA). The categorical variables (i.e., the clinicopathologic features, FGFR3 expression, and FGFR3 mutation) were analyzed by the Chi-square test. Associations between the expression levels of FGFR3 and the OS and DFS of the NSCLC patients were evaluated by a Kaplan-Meier survival analysis with a log-rank test. The association between the risk score and clinical features was detected by univariate and multivariate Cox analyses. The Cox regression model was used to calculate the hazard ratios and 95% confidence intervals. A P value <0.05 was considered statistically significant.
Results
Clinical data
A total of 116 cases of NSCLC were included in the preliminary analysis. Among the 116 cases, the complete clinical records of NSCLC patients were available for 86 cases, and those cases were included in the further analysis. The clinical pathological parameters of the 86 NSCLC patients are summarized in Table 1. There were 57 male patients and 29 female patients in this cohort. The patients had a median age of 58 years (range, 41–80 years) at the time of diagnosis. The clinical follow-up period was up to 64 months. Among the patients, 46 were ever-smoking patients. The cases for AC and SCC were 39 and 47, respectively. The cases for early stage (I–II) and late stage (III) tumors were 59 and 27, respectively. In addition, the cases for the EGFR mutation and wild-type EGFR were 39 and 47, respectively.
The expression of FGFR3 in NSCLC tissues
The FGFR3 expression levels of the 86 NSCLC tissues were evaluated by TMA. FGFR3 expression (both cytoplasmic and membranous) was detected by IHC. The non-expression, low expression, and high expression of FGFR3 in the NSCLC tissues are shown in Figure 1. Based on the IHC results, FGFR3 was immunoreactive in 26 of the 86 NSCLC cases. Further, FGFR3 was positively expressed in 84.6% of the lung AC cases and 15.4% of the lung SCC cases. FGFR3 was positively expressed in 42.3% of the male patients and 57.7% of the female patients. Additionally, FGFR3 was positively expressed in 15.4% of the smoking cases and 84.6% of the non-smoking cases. Further, 69.2% of the early stage tumors (I and II) cases and 30.8% of the late stage tumors (III) cases were positive for FGFR3. Among the FGFR3 positive cases, 80.8% of the cases had the EGFR mutation and 19.2% cases had wild-type EGFR.
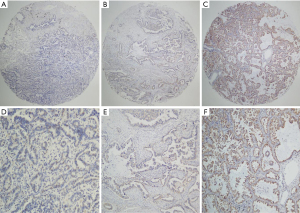
All the samples were further divided into either high or low FGFR3 expression groups. Patients with a smoking history, histology type, T stage, and the EGFR mutation had higher FGFR3 expression rates. However, high FGFR3 expression was not associated with other clinical parameters, such as age, N stage, and clinical stage (see Table 1).
Analysis of FGFR3 mutations in NSCLC samples
DNA was extracted from the paraffin-embedded tissues of 86 NSCLC tissues, and direct sequencing was conducted on exons 7, the extracellular domain, 10 and 15 of FGFR. A total of 72 cases were included in the FGFR3 sequencing analysis. FGFR3 mutations were found in 2/72 (2.8%) NSCLC cases as follows: 1/28 (3.6%) in AC, and 1/44 (2.3%) in SCC. Two cases of the FGFR3 mutation were found in exons 10, but no mutations were found in exons 7 or15 of FGFR3. Among the 70 NSCLC cases with wild-type FGFR3, 20 (28.6%) cases were FGFR3 positive. Due to the small number of FGFR3 mutation cases, the associations between the FGFR fusions and key clinicopathologic characteristics were not assessed.
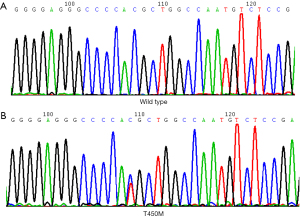
T450M is a novel mutation in FGFR3 TM domain
The mutational analysis of FGFR3 exon 10 showed the only point mutation at T450M. Figure 2 shows heterozygous substitutions C > T at position 1349 in exon 10 of FGFR3, leading to amino acid substitution T450M. The Database Single Nucleotide Polymorphism (dbSNP) indicated that it was not a polymorphism (see Table S1). The mutation was detected in 2 patients. Of whom, 1 was a 54-year-old female, who had undergone a radical lobectomy, and had stage cT2a N1M0 LA. The patient was alive, and the FGFR3 protein expression was positive. The EGFR had 19 deletion mutations. The other patient was a 78-year-old male, who had been diagnosed with SCC. He had undergone radical lobectomy and had stage T2a N1M0. The molecular analysis revealed the negative staining of FGFR3 and wild-type EGFR. T450M in FGFR3 exon 10 was the only mutation detected in the 2 patients. No other mutations in exons 7 and 15 of FGFR3 were detected.
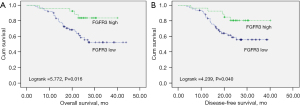
The association between FGFR3 expression and the survival of NSCLC patients
The correlation between FGFR3 expression and the OS and DFS of the NSCLC patients was assessed by a Kaplan-Meier survival analysis. High FGFR3 expression was correlated with better OS (log-rank =5.772, P=0.016; see Figure 3A) and DFS (log-rank =4.239, P=0.040; see Figure 3B).
Univariate and multivariate analyses of the clinical factors and FGFR3 expression associated with the survival of NSCLC patients
Initially, age, smoking, histology type, T stage, N stage, clinical stage, the EGFR mutation, and the expression of FGFR3 were selected for the univariate analysis. The variables were reduced through forward-stepwise regression. The multivariate analysis revealed that FGFR3 was an independent prognostic factor (P=0.024) for the OS of NSCLC patients (see Table 2).
Table 2
| Variable | Subset | HR (95% CI) | P value |
|---|---|---|---|
| Univariate analysis (N=86) | |||
| Age | ≥60 vs. <60 | 0.945 (0.447–1.999) | 0.883 |
| Smoking | Yes vs. no | 1.554 (0.734–3.291) | 0.249 |
| Gender | Male vs. female | 2.501 (0.950–6.582) | 0.063 |
| Histology type | AC vs. SCC | 0.523 (0.236–1.157) | 0.110 |
| T stage | ≤5 vs. >5 cm | 0.827 (0.351–1.950) | 0.664 |
| N stage | 0 vs. 1, 2, 3 | 1.756 (0.830–3.718) | 0.141 |
| Clinical stage | I, II vs. III | 1.303 (0.601–2.826) | 0.503 |
| EGFR mutation | Yes vs. no | 0.523 (0.236–1.157) | 0.110 |
| FGFR3 | Low vs. high | 0.294 (0.102–0.850) | 0.024 |
| Multivariate analysis (N=86) | |||
| FGFR3 | Low vs. high | 0.294 (0.102–0.850) | 0.024 |
FGFR3, fibroblast growth factor receptor 3; NSCLC, non-small cell lung cancer; HR, hazard ratio; CI, confidence interval; AC, adenocarcinoma; SCC, squamous cell carcinoma; EGFR, epidermal growth factor receptor.
Discussion
Clinical trial (NCT01824901) examining FGFR inhibitors in NSCLC is ongoing. New targets for which therapies already exist need to be identified. FGFR3 is a member of the FGFR family, whose alterations may become an effective target for the gene therapy of NSCLC. In this study, the frequencies of protein expression and the FGFR3 mutation in the NSCLC patients were 26/86 (30.2%) and 2/72 (2.8%), respectively. We showed for the first time that FGFR3 is associated with a favorable prognosis and is a novel independent prognostic factor for the OS of NSCLC patients. The high FGFR3 expression group had more females, more non-smokers, more ACs, earlier T stage, fewer lymph node metastases, earlier clinical stage, a higher EGFR mutation rate, and longer OS and DFS. Reasons for the favorable prognosis of NSCLC patients include the small sample size for death events during the follow-up period and the different treatment modalities, which included tyrosine kinase inhibitor (TKI)–treatment and adjuvant chemotherapy. Notably, we discovered that T450M is a novel mutation in the FGFR3 TM domain. T450M in FGFR3 exon 10 was the only mutation detected in 2 patients. No other mutations in exons 7 and 15 of FGFR3 were detected. Further, 2 cases were lung AC with overexpressed FGFR3 and lung SCC with FGFR3 negative staining respectively.
A study reported that the expression rate of FGFR3 was 1.8% (5/275) in lung SCC (26). Conversely, we found that high FGFR3 expression was predominant in lung AC patients, but the positive rate of FGFR3 in lung SCC was low. A previous study has reported that the FGFR3 expression rate is 3.3% (20/612) in NSCLC (27). However, the FGFR3 expression rate was 22.3% (23/103) using the polyclonal antibody from the Abcam company, and 11.7% (12/103) using antibodies from Cell Signaling Technology in 103 lung SCC cases, and the positive expression rate of FGFR3 in NSCLC was not detected. We found a FGRF3-positive expression rate of 15.4% (4/26) using an Abcam monoclonal antibody in lung SCC. There is no unified standard scoring system for FGFR3 expression in NSCLC. However, as FGFR3 in urothelial carcinoma has been widely studied (28) (Table S1), we adopted the scoring system of Cappellen et al. (24). We also reviewed the scoring system for FGFR3 expression from a NSCLC study (29) (Table S2), but the frequency of FGFR3 expression was different in NSCLC. Any discrepancies between previous studies and our findings may be explained by the composition of patients, the heterogeneity of the FGFR3 gene, the cancer stage, antibodies, and the different scoring systems.
Previous studies showed that am EGFR TKI induced FGFR3 signaling by de-repressing FGFR3 expression, and FGFGR3 signaling is a key signaling pathway in the regulation of EGFR TKI resistance (30-32). Thus, de novo FGFR3 high expression may be significantly related to the EGFR mutation state of activation in treatment-naive NSCLC patients. Further, combining EGFR with FGFR specific TKIs may improve the therapeutic effects of EGFR inhibitors.
FGFR3 has been observed to be overexpressed in human hepatocellular carcinoma, oral cancer, and bladder cancer (33). Thus, the overexpression of FGFR3 may play an important role in human carcinogenesis (34). However, little is known about the role of FGFR3 in NSCLC. This study was the first to apply a TMA-based IHC method to examine FGFR3 expression in a cohort of 116 patients. Our results suggest that FGFR3 plays an essential role in NSCLC progression. A recent study showed that miR-100 and FGFR3 protein levels are inversely expressed in the A549 lung cancer cell line (35). However, the mechanisms underlying the upregulation of FGFR3 in A549 cells remain unknown.
High FGFR3 expression is correlated with clinicopathologic factors in lung cancer. Our study showed that FGFR3 was differentially expressed in the lung tissues between the AC and SCC subtypes, which suggests that the tumor behavior may be driven by the morphologic characteristics of tumors (36). The expression of FGFR3 was higher in lung AC than SCC, and higher FGFR3 expression predicted better OS. Additionally, the high FGFR3 expression group had a higher EGFR mutation rate, which suggests that patients in the high FGFR3 expression group received EGFR TKI treatments more than those in the low FGFR3 expression group. Thus, FGFR3 appears to be closely related to AC with the EGFR mutation.
The results of the survival analysis revealed the patients with high FGFR3 expression levels had better OS, which suggests that lower or reduced FGFR3 expression levels may enhance NSCLC progression, while FGFR3 overexpression may attenuate NSCLC progression. FGFR3 overexpression has also been shown to induce the transformation of colon cancer cells (37). FGFR3 upregulation was detected at the messenger ribonucleic acid and protein levels in urinary bladder carcinomas (38). In relation to the role of FGFR3 in the progression of human cancers, some results are contradictory. For example, FGFR3 mutation and expression have been shown to be related to favorable low-grade bladder cancer (39). Conversely, FGFR3 overexpression has also been shown to be a prognostic factor for adverse outcomes for muscle-invasive bladder carcinoma treated with adjuvant chemotherapy (40). FGFR3 expression has also been shown to be associated with a poor prognosis of breast cancer patients (41). Interestingly, in our study, FGFR3 was associated with a favorable prognosis in patients with NSCLC and served as an independent prognostic factor for the OS of NSCLC patients. There are several explanations regarding the contradictory findings among different studies. In the present study, we only included the patients with surgically resectable NSCLC at the I–IIIA stage, and the favorable prognostic role of FGFR3 may only occurred in patients with early-stage NSCLC. This study is a retrospective study, the study is based on the 7th edition of TNM staging classification. However, the current NSCLC staging has been updated to the 8th edition, this staging classification may affect patient management, because many clinicians make management based on stage Therefore, the staging system of this study may affect the judgment of prognostic results and have a certain impact on the treatment plan. Kaplan-Meier survival analysis showed that patients with high FGFR3 expression had significantly improved OS and significantly prolonged DFS compared with patients with low FGFR3 expression. It may be because the expression of FGFR3 in lung adenocarcinoma is higher than that in lung squamous cell carcinoma, and the EGFR mutation rate in the group with high FGFR3 expression is higher, and there is a chance to receive EGFR-TKI treatment in the future. Therefore, it is speculated that patients with high FGFR3 expression have longer OS and DFS, and are closely associated with higher EGFR mutations. However, the mechanistic actions still need to be further investigated.
Our study also found no correlation between protein expression and the FGFR3 mutation. FGFR3 mutations have been described previously in urothelial carcinoma (42,43). However, similar to a previous report (44), the frequency of the FGFR3 mutation in the NSCLC patients in our cohort was low. Studies have reported that FGFR3 aberrations in NSCLC occurred in the FGFR3 fusion gene type (27) (Table S3). Pros et al. did not discover any FGFR3 mutation in 50 cases of NSCLC (45). Shinmura et al. reported that the FGFR3 mutation rate was 0.9% (2/214) in NSCLC (38), which was lower than the rate reported in the present study (Table S4). However, similar to the results of our study, the frequency rate in lung SCC was reported to be 3.2% (2/63). We found that FGFR3 mutations were not associated with the prognosis of NSCLC patients, but this may be due to the limited sample size.
This study had several limitations. First, as a single-center retrospective study, selection bias was unavoidable. Second, IHC and Sanger sequencing were combined to detect FGFR3 status, and the frequency of FGFR3 alterations in China differ to those of other countries. Third, currently, there are no consistent standards, including antibody and scoring system standards. Fourth, this was only a preliminary study, and more thorough studies using human cells or samples need to be conducted to provide further insights into the function of FGFR3 in NSCLC and the progression of NSCLC.
Conclusions
FGFR3 alterations, including expression and mutation alterations, in NSCLC could be promising therapeutic targets. We found that the frequency of FGFR3 expression in NSCLC, especially AC, is high, but the FGFR3 mutation rate is low and is not correlated with the histology type. FGFR3 may serve as an independent prognostic factor for the OS of NSCLC patients.
Acknowledgments
Funding: This work was supported by grants from the Guangzhou Health Science and Technology Project (Nos. 20202A010014, 20211A011091, and 20212A011024), the Traditional Chinese Medicine Bureau Research Project of Guangdong Province in China (No. 20202111), and the Science and Technology Program of Guangzhou Health Commission (Nos. 20211A011088, 20222A010047).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1523/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1523/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1523/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Guangzhou Medical University (No. 50, 2019), and informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med 2019;27:167-70.
- Chen P, Liu Y, Wen Y, et al. Non-small cell lung cancer in China. Cancer Commun (Lond) 2022.
- Yu P, He X, Lu F, et al. Research progress regarding long-chain non-coding RNA in lung cancer: a narrative review. J Thorac Dis 2022;14:3016-29. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Chen W, Li L, Cheng S, et al. The Efficacy of Immune Checkpoint Inhibitors vs. Chemotherapy for KRAS-Mutant or EGFR-Mutant Non-Small-Cell Lung Cancers: A Meta-Analysis Based on Randomized Controlled Trials. Dis Markers 2022;2022:2631852. [Crossref] [PubMed]
- Hao Y, Zhang X, Yu L. Immune checkpoint inhibitor-related pneumonitis in non-small cell lung cancer: A review. Front Oncol 2022;12:911906. [Crossref] [PubMed]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [Crossref] [PubMed]
- Remon J, Steuer CE, Ramalingam SS, et al. Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann Oncol 2018;29:i20-i7. [Crossref] [PubMed]
- Tan CS, Kumarakulasinghe NB, Huang YQ, et al. Third generation EGFR TKIs: current data and future directions. Mol Cancer 2018;17:29. [Crossref] [PubMed]
- Tan AC, Pavlakis N. Anti-Angiogenic Therapy in ALK Rearranged Non-Small Cell Lung Cancer (NSCLC). Int J Mol Sci 2022;23:8863. [Crossref] [PubMed]
- Gaughan EM, Costa DB. Genotype-driven therapies for non-small cell lung cancer: focus on EGFR, KRAS and ALK gene abnormalities. Ther Adv Med Oncol 2011;3:113-25. [Crossref] [PubMed]
- Ramalingam SS, Owonikoko TK, Khuri FR. lung cancer: new biological insights and recent therapeutic advances. CA Cancer J Clin 2011;61:91-112. [Crossref] [PubMed]
- Chew NJ, Nguyen EV, Su SP, et al. FGFR3 signaling and function in triple negative breast cancer. Cell Commun Signal 2020;18:13. [Crossref] [PubMed]
- Sakashita T, Yanagitani N, Koike S, et al. Fibroblast growth factor receptor 3 overexpression mediates ALK inhibitor resistance in ALK-rearranged non-small cell lung cancer. Cancer Sci 2022;113:3888-900. [Crossref] [PubMed]
- Liu C, Liu C, Liao J, et al. Genetic correlation of crizotinib efficacy and resistance in ALK- rearranged non-small-cell lung cancer. Lung Cancer 2022;171:18-25. [Crossref] [PubMed]
- Hart KC, Robertson SC, Donoghue DJ. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, Stat activation, and phosphatidylinositol 3-kinase activation. Mol biol cell 2001;12:931-42. [Crossref] [PubMed]
- L'Hôte CG, Knowles MA. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Exp cell res 2005;304:417-31. [Crossref] [PubMed]
- Chellaiah AT, McEwen DG, Werner S, et al. fibroblast growth factor receptor (fgfr) 3. alternative splicing in immunoglobulin-like domain iii creates a receptor highly specific for acidic fgf/fgf-1. J Biol Chem 1994;269:11620-7.
- Murgue B, Tsunekawa S, Rosenberg I, et al. identification of a novel variant form of fibroblast growth factor receptor 3 (fgfr3 iiib) in human colonic epithelium. Cancer Res 1994;54:5206-11.
- Sturla LM, Merrick AE, Burchill SA. fgfr3iiis: a novel soluble fgfr3 spliced variant that modulates growth is frequently expressed in tumour cells. Br J Cancer 2003;89:1276-84. [Crossref] [PubMed]
- Acquaviva J, He S, Zhang C, et al. FGFR3 translocations in bladder cancer: differential sensitivity to HSP90 inhibition based on drug metabolism. Mol Cancer Res 2014;12:1042-54. [Crossref] [PubMed]
- Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet 1999;23:18-20. [Crossref] [PubMed]
- Bodoor K, Ghabkari A, Jaradat Z, et al. FGFR3 mutational status and protein expression in patients with bladder cancer in a Jordanian population. Cancer epidemiol 2010;34:724-32. [Crossref] [PubMed]
- Kompier LC, Lurkin I, van der Aa MN, et al. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One 2010;5:e13821. [Crossref] [PubMed]
- Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol 2013;230:270-6. [Crossref] [PubMed]
- Nakanishi Y, Akiyama N, Tsukaguchi T, et al. Mechanism of Oncogenic Signal Activation by the Novel Fusion Kinase FGFR3-BAIAP2L1. Mol Cancer Ther 2015;14:704-12. [Crossref] [PubMed]
- Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Parker BC, Engels M, Annala M, et al. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J Pathol 2014;232:4-15. [Crossref] [PubMed]
- Chen L, Zhang Y, Yin L, et al. Fibroblast growth factor receptor fusions in cancer: opportunities and challenges. J Exp Clin Cancer Res 2021;40:345. [Crossref] [PubMed]
- Carotenuto M, Sacco A, Forgione L, et al. Genomic alterations in cholangiocarcinoma: clinical significance and relevance to therapy. Explor Target Antitumor Ther 2022;3:200-23. [Crossref] [PubMed]
- Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231-5. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795-803. [Crossref] [PubMed]
- Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636-47. [Crossref] [PubMed]
- Liao RG, Jung J, Tchaicha J, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res 2013;73:5195-205. [Crossref] [PubMed]
- Shinmura K, Kato H, Matsuura S, et al. A novel somatic FGFR3 mutation in primary lung cancer. Oncol Rep 2014;31:1219-24. [Crossref] [PubMed]
- Kim Y, Hammerman PS, Kim J, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J clin oncol 2014;32:121-8. [Crossref] [PubMed]
- Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin cancer res 2014;20:4107-14. [Crossref] [PubMed]
- Tsim S, O'Dowd CA, Milroy R, et al. staging of non-small cell lung cancer (nsclc): a review. Respir Med 2010;104:1767-74. [Crossref] [PubMed]
- Neuzillet Y, van Rhijn BW, Prigoda NL, et al. FGFR3 mutations, but not FGFR3 expression and FGFR3 copy-number variations, are associated with favourable non-muscle invasive bladder cancer. Virchows arch 2014;465:207-13. [Crossref] [PubMed]
- Bernardo P, Budetta M, Aliberti F, et al. Temporal lobe malformations, focal epilepsy, and FGFR3 mutations: a non-causal association? Neurol Sci 2021;42:2063-7. [Crossref] [PubMed]
- Turo R, Harnden P, Thygesen H, et al. FGFR3 expression in primary invasive bladder cancers and matched lymph node metastases. J Urol 2015;193:325-30. [Crossref] [PubMed]
- Pros E, Lantuejoul S, Sanchez-Verde L, et al. Determining the profiles and parameters for gene amplification testing of growth factor receptors in lung cancer. Int J Cancer 2013;133:898-907. [Crossref] [PubMed]


