
Deletion of LCMR1 in alveolar type II cells induces lethal impairment of lung structure and function in adult mice
Highlight box
Key findings
• Deletion of LCMR1 specifically in AT2 cells results in impaired lung structure and function that is lethal to adult mice, which highlights the importance of LCMR1 in the maintenance of AT2 cell homeostasis.
What is known and what is new?
• LCMR1 is related to multiple biological processes in different cancer cells in vitro; however, its role in normal tissues and cells has not been fully elucidated.
• LCMR1 is critical for the maintenance of AT2 cell homeostasis.
What are the implications, and what should change now?
• This is the first reported AT2 cell-specific LCMR1 conditional knockout mouse model to date, which can provide a valuable reference for investigating the function of LCMR1 in other tissues or cells.
Introduction
Lung cancer metastasis-related protein 1 (LCMR1) was originally identified by differential display polymerase chain reaction (PCR) between human large cell lung carcinoma cell lines 95C and 95D by our team in 2002 (Genbank accession number: AY148462) (1). Our previous work showed that LCMR1 is strongly overexpressed in non-small cell lung cancer and its expression is significantly associated with the clinical stage of patients (1). In addition, the knockdown of LCMR1 was reported to induce cell apoptosis in different cancer cells in vitro (2-4). LCMR1 is also known as a subunit of the Mediator complex, called MED19 (5). The Mediator is an evolutionarily conserved multisubunit protein complex that acts as a critical transcription cofactor conveying information between gene-specific transcription factors and RNA polymerase II general transcription machinery (6,7). The mammalian Mediator comprises up to 30 distinct subunits with unique biological functions (8,9). LCMR1 is a crucial facilitator of peroxisome proliferator-activated receptor gamma (PPARγ)-mediated gene expression that is required for adipogenesis and maintenance of white adipose tissue (WAT) mass in mice (10). LCMR1 also acts as a GATA cofactor and MED1 interactor in Drosophila (11). Additionally, LCMR1 and MED26 are synergistic functional targets of the RE1 silencing transcription factor (REST) in the epigenetic silencing of neuronal gene expression in non-neuronal cells (12). However, LCMR1 is still a relatively poorly characterized gene and the role of LCMR1 in normal lung tissue remains to be elucidated.
The lung is composed of air-conducting bronchi terminating in air-exchanging alveoli. The alveolar epithelium consists of squamous alveolar type I cells (AT1 cells), which are responsible for gas exchange, and cuboidal alveolar type II cells (AT2 cells), which serve as the only source of pulmonary surfactant, reducing surface tension and preventing lung collapse at end-expiration. AT2 cells also participate in modulation of immune responses and transportation of ions and fluids. In addition, AT2 cells have been recognized as alveolar stem cells with the capacity for self-renewal and differentiation into AT1 cells during lung homeostasis and injury repair (13,14). Specific subgroups of AT2 cells execute the progenitor role in repairing the alveoli, which preferentially re-enter the cell cycle, self-renew and regenerate mature AT1 and AT2 cells after acute lung injury (15,16). However, damaged or denuded alveolar epithelium often triggers defective or overwhelmed repair processes (17).
In this study, we generated a novel tamoxifen-inducible, AT2 cell-specific LCMR1 conditional knockout mouse model by breeding mice carrying floxed LCMR1 alleles with Sftpc-CreERT2 mice, aiming to investigate the effects of AT2 cell-specific LCMR1 deletion on lung structure and function in adult mice. We demonstrated that AT2 cell-specific LCMR1 deletion caused increased weight loss, damaged lung structure, impaired lung function, and reduced survival of mice. The lethal phenotype we observed may result from severe AT2 cell depletion, pulmonary surfactant dysfunction, and alveolar structure destruction. These results indicate that LCMR1 may play an important role in the maintenance of AT2 cell homeostasis. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-293/rc).
Methods
Mice
Mice carrying the floxed LCMR1 allele were generated in cooperation with Cyagen Biosciences Inc. (Suzhou, China) by combining CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) and the Cre/loxP system. Five exons of the LCMR1 gene have been identified with the ATG start codon in exon 1 and the TAA stop codon in exon 5. Exons 2–4 were selected as the conditional knockout region. Briefly, to engineer the target vector, homology arms and a conditional knockout region were generated by PCR using bacterial artificial chromosome (BAC) clones RP24-220L15 and RP23-427H11 from the C57BL/6J library as templates. The donor vector containing loxP sites, guide RNAs (gRNA1: TTTAAAGGAATTCCACTCCCTGG; gRNA2: TTCACTGGGTTGTTCTAGGCAGG), and Cas9 messenger RNA (mRNA) were co-injected into fertilized mouse eggs to generate targeted conditional knockout mice.
Potential founders were identified by PCR and Sanger sequencing employing mouse tail DNA (data not shown) and then bred to WT mice. F1 offspring (LCMR1flox/wt) were also identified by PCR with specific primers for the 5'-loxP site (F-CACGTAAACGGCCACAAGTTCGAG, R-CATGAGGTTAAGGAGAGTTGGGAC) and 3'-loxP site (F-GGTAACTAGGGAGGTCAGAGACATGAG, R-CGAAGTTATGTGCACAGTACTGTGG), respectively, and further confirmed via Sanger sequencing and Southern blot analysis (detailed methods not shown).
Sftpc-CreERT2 mice (Jackson Laboratory, Bar Harbor, ME, USA) express a tamoxifen-inducible Cre recombinase controlled by the Sftpc promoter, which can mediate the deletion of targeted sequences flanked by the loxP sites specifically in AT2 cells. LCMR1flox/wt mice were mated to Sftpc-CreERT2 mice, and the offspring were screened for the presence of floxed LCMR1 and the Sftpc-CreERT2 transgene. The double heterozygous mice were then intercrossed to obtain Sftpc-CreER+/−; LCMR1flox/flox mice, which were used as breeding pairs to generate AT2 cell-specific LCMR1 conditional knockout mice (Sftpc-CreERT2; LCMR1flox/flox) and littermate control mice (LCMR1flox/flox) for the following phenotype study.
Tamoxifen (20 mg/mL in corn oil; Sigma, T5648, St. Louis, MO, USA) was administered via intraperitoneal injection (50 mg/kg) to 6-week-old mice for 5 consecutive days, after which the Sftpc-CreERT2; LCMR1flox/flox mice were termed LCMR1ΔAT2 mice. The control littermates (LCMR1flox/flox) received the same doses of tamoxifen. The day of the last dose of tamoxifen administration was recorded as day 0. The mice were housed in individually ventilated cages with free access to food and water under specific pathogen-free (SPF) conditions. All mice were maintained in the C57BL/6J background. A protocol was prepared before the study without registration. All animal experiments were approved by the Institutional Animal Care Use Committee of Chinese PLA General Hospital (No. 2018-X14-25), in compliance with institutional guidelines for the care and use of animals.
Genotyping
Mouse tail genomic DNA (gDNA) was extracted using the DNA extraction kit (Takara, 9765, Shiga, Japan) according to the manufacturer’s protocol. The presence of the two loxP sites and the Sftpc-CreERT2 transgene was determined by PCR with primers shown in Table S1. DNA of the sorted AT2 cells from lung tissue was also isolated to confirm LCMR1 deletion through PCR after tamoxifen administration. PCR was performed in a 50 µL reaction volume using Premix Taq (TaKaRa, RR902A) on a T100™ Thermal Cycler (Bio-Rad, Hercules, CA, USA), and PCR products were detected by agarose gel electrophoresis and visualized using a Molecular Imager ChemiDoc XR System (Bio-Rad) with Image Lab software, version 3.0 (Bio-Rad). The expected sizes of PCR products were also shown in Table S1.
Histopathological analysis
The lung tissues of the mice were fixed in 4% paraformaldehyde for 24 h at 4 ℃, dehydrated in a graded ethanol series, and then embedded in paraffin. The paraffin-embedded block was cut into 5-µm sections which were then deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E) reagents (Soonbio, Beijing, China) according to the standard procedure. Collagen fibers in the lung sections were detected by Masson’s trichrome staining and Sirius red staining using standard procedures. The stained lung sections were examined under a light microscope (Olympus, Tokyo, Japan).
Immunofluorescence and TUNEL staining
For immunofluorescence staining, the 5-µm paraffin-embedded sections were deparaffinized and rehydrated, followed by heat-mediated antigen retrieval. The sections were blocked with 3% bovine serum albumin (BSA) for 30 min at room temperature and incubated with primary antibodies overnight at 4 ℃ including anti-prosurfactant protein C (proSP-C) (Millipore, AB3786, Temecula, CA, USA) and anti-α-smooth muscle actin (α-SMA) (Proteintech, 14395-1-AP, Wuhan, China). The sections were then incubated with a secondary antibody (Cy3-conjugated goat anti-rabbit IgG) for 1 h at room temperature. The nuclei were then stained with 4’,6-diamidino-2-phenylindole (DAPI) for 10 min. The apoptosis of AT2 cells within lung sections was analyzed using proSP-C and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) co-staining, along with double immunofluorescence staining for proSP-C and cleaved caspase-3 (Servicebio, GB11532, Wuhan, China). Images were captured using an Olympus IX81 fluorescence microscope and a Nikon Eclipse Ti confocal microscope (Tokyo, Japan).
Transmission electron microscopy
Lung tissues were dissected, minced to approximately 1 mm3 pieces as soon as possible, and fixed overnight at 4 ℃ using 2.5% glutaraldehyde. The samples were then postfixed with 1% osmium tetroxide in cacodylate buffer, dehydrated in a graded ethanol series, and embedded in epoxy resin. Ultrathin sections (70 nm) were obtained using an Ultracut E ultramicrotome (Leica, Wetzlar, Germany), and stained with uranyl acetate and lead citrate. The sections were subsequently examined under an H-7650 transmission electron microscope (Hitachi, Tokyo, Japan) with an attached image capture software.
Lung dissociation and AT2 cells isolation
Lung dissociation was performed as described previously with simple modification (18). The protease solution used for lung digestion in this study contained Collagenase Type I (450 U/mL; Sigma, C0130), Elastase (4 U/mL; Sigma, E1250), Dispase (5 U/mL; Sigma, D4693), and DNaseI (0.33 U/mL; Roche, 10104159001, Mannheim, Germany) dissolved in Dulbecco’s minimal essential medium/F12 (DMEM/F12) (Gibco, 11330021, Grand Island, NY, USA). The whole-lung suspension was blocked with TruStain FcX anti-mouse CD16/CD32 (BioLegend, 101319, San Diego, CA, USA) in the Dulbecco’s phosphate buffered saline (D-PBS) buffer with 10% fetal bovine serum (FBS), and then stained with anti-CD31-FITC (eBioscience, 11-0311-81), anti-CD45-FITC (eBioscience, 11-0451-81), and anti-CD326-APC (eBioscience, 17-5791-82). Fluorescence activated cell sorting (FACS) was performed on an SH800S Cell Sorter (Sony, San Jose, CA, USA).
Quantitative RT-PCR
Total RNA was isolated using RNASimple Total RNA Kit (Tiangen, DP451, Beijing, China) according to the manufacturer’s instructions. Complementary DNA (cDNA)was synthesized using the PrimeScript RT reagent Kit with a gDNA Eraser (Takara, RR047A). Quantitative RT-PCR (qPCR) was performed in a 20 µL reaction volume using KAPA SYBR FAST Universal (KAPA Biosystems, KK4601, Cape Town, South Africa) on MyiQ2 Two-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Primers used for amplification were listed in Table S2. The relative mRNA expression was assessed using the standard 2−ΔΔCt method normalized by β-actin.
Western blot
The total protein were extracted using Radioimmunoprecipitation Assay (RIPA) lysis buffer supplemented with protease and phosphatase inhibitor cocktails (Applygen, C1055, Beijing, China). Protein concentration was measured with a bicinchoninic acid (BCA) protein assay kit (Applygen, P1511). Protein was separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Immunoblotting was performed using specific antibodies, including anti-MED19 (Invitrogen, PA5-78656, Carlsbad, CA, USA), anti-α-SMA (Proteintech), anti-SP-A (Proteintech, 11850-1-AP), anti-SP-B (Bioss, bs-22341R, Beijing, China), anti-SP-C (Proteintech, 10774-1-AP), anti-SP-D (Proteintech, 11839-1-AP), anti-P53 (CST, 2524S, Danvers, MA, USA), anti-B-cell lymphoma-2 (Bcl-2) (PTM Bio., PTM-5777, Hangzhou, China), anti-β-actin (Proteintech, 20536-1-AP), and anti-α-tubulin (PTM Bio., PTM-5442). The protein bands were visualized with a Chemiluminescent HRP Substrate (Millipore) using an automatic chemiluminescence imaging system (Tanon, Shanghai, China). The band intensity was quantified using ImageJ software, version 1.52 (National Institutes of Health) and normalized to α-tubulin or β-actin.
Analysis of bronchoalveolar lavage fluid
After the mice were anesthetized, their tracheas were cannulated and their lungs were lavaged three times with 1 mL of sterile saline. The recovered bronchoalveolar lavage fluid (BALF) was then centrifuged at 500 g for 10 min at 4 ℃. The supernatant was used to measure the total protein concentration using a BCA Protein Assay kit (Applygen) and cytokine levels using a LEGENDplex Mouse Inflammation Panel (BioLegend, 740446). The cell pellet was resuspended to determine the total cell numbers using a hemocytometer. Differential cell counts were determined manually from the cytospins of BALF cell pellets stained with H&E, including neutrophils, macrophages, and lymphocytes.
Lung wet/dry weight ratio
After the mice were anesthetized, the right lung was removed and weighed to obtain the wet weight. Subsequently, the samples were desiccated in an oven at 80 ℃ for 48 h and re-weighed to determine the dry weight. The lung wet/dry weight ratio was then calculated.
Pulmonary function measurement
Pulmonary function was measured using the AniRes2005 lung function testing system (Bestlab, Beijing, China). Briefly, the mice were anesthetized, cannulated via tracheotomy, placed into a sealed plethysmograph chamber, and then connected to a computer-controlled, mechanically ventilated system (AniRes2005). Five consecutive measurements were collected for each mouse, and the average value was used for further analyses of the forced vital capacity (FVC), dynamic lung compliance (Cdyn), expiratory resistance (Re), and inspiratory resistance (Ri).
Statistical analysis
Data were exhibited as the mean ± standard error of mean (SEM). Statistical analyses were performed using GraphPad Prism 5.0 software (National Institutes of Health). Comparisons between two groups were processed using the unpaired Student’s t-test. P<0.05 was considered statistically significant.
Results
Generation of AT2 cell-specific LCMR1 conditional knockout mice
To explore the role of LCMR1 in lung tissue, we generated a tamoxifen-inducible, AT2 cell-specific LCMR1 conditional knockout mouse model (Sftpc-CreERT2; LCMR1flox/flox) for which the gene targeting strategy was briefly illustrated in Figure 1A. Initially, we constructed a conditional knockout mouse line with exons 2–4 of the LCMR1 gene flanked by two loxP sites (LCMR1flox/wt), which was identified and verified by PCR, Sanger sequencing, and Southern blot analysis (Figure S1A-S1C). To obtain the Sftpc-CreERT2; LCMR1flox/flox mice for AT2 cell-specific LCMR1 deletion, a subsequent breeding scheme was simply depicted in Figure 1B. The genotypic distribution of offspring was in accordance with Mendel’s law of inheritance (data not shown, only the genotype that we needed for further breeding is listed in the schematic diagram).
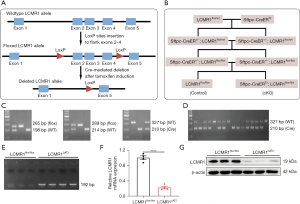
We first crossed LCMR1flox/wt mice with Sftpc-CreERT2 mice to obtain Sftpc-CreER+/−; LCMR1flox/wt mice, which were further bred for Sftpc-CreER+/−; LCMR1flox/flox mice (Figure 1C). Next, the Sftpc-CreER+/−; LCMR1flox/flox mice were used as breeding pairs to further generate Sftpc-CreERT2; LCMR1flox/flox mice together with LCMR1flox/flox mice (Sftpc-CreERT2 negative) as the littermate control. Based on Mendel’s law of inheritance, the offspring were all expected to harbor the homozygous floxed LCMR1 allele, and thus, genotyping was performed to detect the presence of the Sftpc-CreERT2 transgene (Figure 1D). Once activated by tamoxifen administration, Cre recombinase can mediate LCMR1 deletion specifically in the AT2 cells of Sftpc-CreERT2; LCMR1flox/flox. Six-week-old mice were intraperitoneally injected with tamoxifen for 5 consecutive days, after which Sftpc-CreERT2; LCMR1flox/flox mice were termed LCMR1ΔAT2 mice. The DNA of AT2 cells sorted on day 7 was extracted, and LCMR1 deletion was confirmed by PCR with a 192-bp band detected only in LCMR1ΔAT2 mice (Figure 1E). The analysis of LCMR1 mRNA expression by qPCR demonstrated a dramatic reduction in the AT2 cells of LCMR1ΔAT2 mice compared with LCMR1flox/flox mice (Figure 1F). Western blot analysis showed that the LCMR1 protein expression was also significantly reduced in the AT2 cell of LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice (Figure 1G). These results further verified that we successfully generated a tamoxifen-inducible, AT2 cell-specific LCMR1 conditional knockout mouse model.
Effects of AT2 cell-specific LCMR1 deletion on lung structure and permeability
The histopathology of lungs from LCMR1ΔAT2 and LCMR1flox/flox mice was examined to assess the effect of LCMR1 deletion on lung structure. H&E staining showed prominent structure damage in the lung sections of LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice on day 14 after tamoxifen administration (Figure 2A). Specifically, the AT2 cell-specific deletion of LCMR1 resulted in inflammatory cell infiltration, hemorrhage, edema, and alveolar wall thickening in the lung sections of LCMR1ΔAT2 mice, which were rarely found in LCMR1flox/flox mice. There was almost no normal alveolar structure in the areas with severe pathological damage in LCMR1ΔAT2 mice.
We further examined the ultrastructure of lung tissues through transmission electron microscopy (TEM). Consistent with the histological analysis, the TEM results demonstrated that the alveolar structure was disordered in LCMR1ΔAT2 mice, while LCMR1flox/flox mice exhibited a mostly normal structure (Figure 2B). The air-blood barrier was thicker in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice. Additionally, there were dysmorphic or electron-dense lamellar bodies in AT2 cells of LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice.
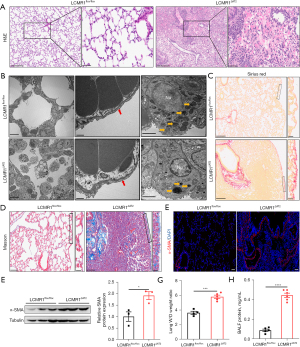
Sirius red staining of lung sections revealed enhanced collagen deposition mainly around the bronchioles or adjacent to the pleura in LCMR1ΔAT2 mice compared with LCMR1flox/flox mice (Figure 2C). Collagen deposition was similarly evidenced by Masson’s trichrome staining (Figure 2D). Furthermore, we also detected the expression of α-smooth muscle actin-positive (α-SMA), a marker of the myofibroblasts (19). The immunofluorescence staining results showed more α-smooth muscle actin-positive (α-SMA-positive) cells in the lung sections of LCMR1ΔAT2 mice compared with those in LCMR1flox/flox mice (Figure 2E). Additionally, western blot showed a significant increase in the whole-lung protein expression of α-SMA in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice (Figure 2F). Besides the pathological changes, we found that the lung wet/dry weight ratio was significantly higher in LCMR1ΔAT2 mice than that in LCMR1flox/flox mice (Figure 2G), accompanied by a significant increase in BALF protein concentration collected from LCMR1ΔAT2 mice compared with that from LCMR1flox/flox mice (Figure 2H), which was indicative of higher lung permeability.
Effects of AT2 cell-specific LCMR1 deletion on body weight, pulmonary function, and survival
We monitored the health status of LCMR1ΔAT2 mice and LCMR1flox/flox mice daily after tamoxifen administration. Data on body weight changes were expressed as a percentage of body weight at day 0 (Figure 3A). LCMR1ΔAT2 mice exhibited increased body weight loss as early as day 4 after tamoxifen administration, whereas all LCMR1flox/flox mice were healthy and continued to gain weight throughout the observation period. There was a significant difference in the weight change of LCMR1ΔAT2 mice and LCMR1flox/flox mice after day 6, which increased with time. Despite prominent body weight loss, LCMR1ΔAT2 mice exhibited shortness of breath, a hunched posture, and decreased activity, followed by death. Notably, the LCMR1ΔAT2 mice showed significantly increased mortality compared with the LCMR1flox/flox mice (Figure 3B). The survival of LCMR1ΔAT2 mice was significantly decreased with the onset of mortality as early as day 10 after tamoxifen administration. There were no surviving LCMR1ΔAT2 mice after day 17, with a median survival of 14 days post-tamoxifen, whereas all LCMR1flox/flox mice were alive and healthy throughout the 30 days observed.
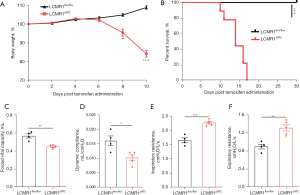
We also examined whether the observed histopathological changes have any effect on lung function through invasive pulmonary function testing. The forced vital capacity (FVC) was significantly decreased in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice (Figure 3C). Additionally, the dynamic compliance (Cdyn) was significantly lower in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice (Figure 3D). We also found increases in the inspiratory resistance (Ri) and expiratory resistance (Re) of LCMR1ΔAT2 mice compared with that of LCMR1flox/flox mice (Figure 3E,3F). These data suggested increased airway resistance and decreased lung compacity and compliance in LCMR1ΔAT2 mice.
Effects of AT2 cell-specific LCMR1 deletion on cellularity and inflammatory cytokines in BALF
To evaluate the effect of LCMR1 deletion on the inflammatory response in the lung, we detected the total cell counts, differential cell counts, and typical cytokine levels in BALF collected from LCMR1flox/flox and LCMR1ΔAT2 mice on day 14 after tamoxifen administration. BALF from LCMR1ΔAT2 mice exhibited a substantial increase in total cell counts compared with that from LCMR1flox/flox mice (Figure 4A). We further performed differential cell counts, which revealed significantly increased neutrophils from LCMR1ΔAT2 mice compared with LCMR1flox/flox mice (Figure 4B). Similar trends were also observed in the numbers of macrophages and lymphocytes in BALF (Figure 4C,4D).
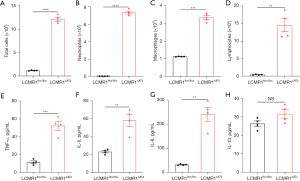
Regarding typical cytokines, the concentrations of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-6 and IL-10 were also detected in BALF from LCMR1flox/flox and LCMR1ΔAT2 mice on day 14 after tamoxifen administration. We also observed significantly elevated levels of proinflammatory cytokines in BALF from LCMR1ΔAT2 mice compared with that from LCMR1flox/flox mice, including TNF-α, IL-1β, and IL-6 (Figure 4E-4G). The level of the typical anti-inflammatory cytokine, IL-10, also trended higher in LCMR1ΔAT2 mice compared with LCMR1flox/flox mice but the increase was not significantly different between the two groups (Figure 4H). Alterations of cell and cytokine profiles in BALF indicated an enhanced inflammatory response in the lungs of LCMR1ΔAT2 mice, which was consistent with the histological changes.
Effects of LCMR1 deletion on the AT2 cell number and pulmonary surfactants
Next, we assessed the impact of LCMR1 on the number and function of AT2 cells. Immunofluorescence staining for the AT2 cell marker proSP-C (20) was performed on lung sections from LCMR1flox/flox and LCMR1ΔAT2 mice on day 14 after tamoxifen administration, which revealed an apparent reduction of proSP-C-positive (proSP-C+) cells in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice, indicating a loss of AT2 cells (Figure 5A). As AT2 cells synthesize and secrete surfactant proteins, we then analyzed the expressions of SP-A, SP-B, SP-C, and SP-D in lung tissue by qPCR and western blot analysis. qPCR analysis showed a significant reduction in the mRNA expressions of SP-A, SP-B, and SP-C in the lung tissues of LCMR1ΔAT2 mice compared with those of LCMR1flox/flox mice, while there was no significant change in whole-lung SP-D mRNA expression in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice (Figure 5B-5E).
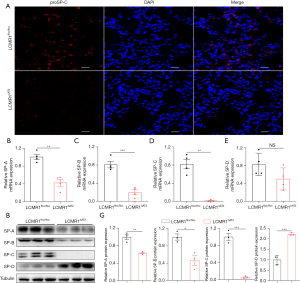
Similarly, western blot analysis revealed decreased protein levels of SP-A and SP-B, as well as SP-C which was nearly undetectable in the lung tissues of LCMR1ΔAT2 mice compared with that of LCMR1flox/flox mice (Figure 5F,5G). On the contrary, the whole-lung SP-D protein level was significantly increased in LCMR1ΔAT2 mice compared with that in LCMR1flox/flox mice (Figure 5F,5G). These results indicated that LCMR1 deficiency led to the loss of AT2 cells and the dysfunction of pulmonary surfactants.
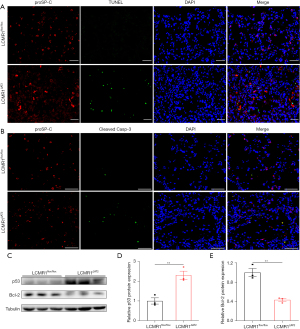
AT2 cell-specific LCMR1 deletion promotes apoptosis in AT2 cells
Since LCMR1 knockdown was reported to promote apoptosis in lung cancer cells (2), we hypothesized that apoptosis may also play an important role in the loss of AT2 cells in LCMR1ΔAT2 mice. To detect AT2 cell apoptosis, we performed double staining for proSP-C and TUNEL on lung sections from LCMR1flox/flox and LCMR1ΔAT2 mice on day 10 after tamoxifen administration (Figure 6A), which showed more proSP-C and TUNEL double positive (proSP-C+TUNEL+) cells in LCMR1ΔAT2 mice compared with those in LCMR1flox/flox mice. Likewise, double staining for proSP-C and cleaved caspase-3 on lung sections also showed more apoptotic AT2 cells in LCMR1ΔAT2 mice than those in LCMR1flox/flox mice (Figure 6B). Additionally, elevated apoptosis in AT2 cells was further confirmed by western blot analysis for p53 and Bcl-2 protein expression, which exhibited upregulated p53 expression paralleled by downregulated Bcl-2 expression in AT2 cells from LCMR1ΔAT2 mice compared with that from LCMR1flox/flox mice (Figure 6C-6E).
Discussion
As LCMR1 was first identified from human large-cell lung carcinoma cell lines, previous studies of LCMR1 mainly focused on its relationship with cancer, especially regarding cell growth, apoptosis, invasion, and metastasis in different cancer types (2-4,21-25). LCMR1 down-regulation can induce laryngocarcinoma cancer cell apoptosis via activation of apoptotic protease activating factor 1 (Apaf-1) (4). LCMR1 can also promote melanoma cell invasion through activating Tetraspanin 8 (Tspan8) protein expression (21). Additionally, LCMR1 can promote breast cancer progression by regulating the EGFR/MEK/ERK signaling pathway (22). Simultaneously, given its role as a subunit of the Mediator complex, LCMR1 is also expected to function as a co-activator of RNA polymerase II enzyme to regulate gene transcription in normal tissues and cells, which has rarely been reported. In this study, we generated the first tamoxifen-inducible, AT2 cell-specific LCMR1 conditional knockout mouse model and preliminarily characterized its phenotype. We found prominent structural abnormalities in the lung tissues of LCMR1ΔAT2 mice, including inflammatory cell infiltration, collagen deposition, alveolar hemorrhage, and edema. Consistently, we also observed impaired pulmonary function, including increased airway resistance and decreased lung compacity and compliance, in LCMR1ΔAT2 mice. All LCMR1ΔAT2 mice died within 17 days after tamoxifen administration. Collectively, these results showed that AT2 cell-specific LCMR1 deletion could induce lethal lung structure and function alterations in adult mice.
CRISPR/Cas9 gene editing technology can effectively induce precise cleavage at target sites in the genome based on short RNA-guided DNA recognition (26,27). Due to its high efficiency and precision, CRISPR/Cas9 has been used to manipulate genes in vitro and in vivo to investigate phenotypic changes and underlying mechanisms of respiratory diseases (28,29). CRISPR/Cas9 can also be used for drug screening, target validation and novel target discoveries, especially in lung cancer (30,31). The Cre/loxP system can efficiently act to direct tissue-specific, site-specific, and heritable chromosomal DNA recombination events in transgenic mice (32). Therefore, combining CRISPR/Cas9 and the Cre/loxP system makes it possible to precisely investigate gene functions in specific cells or tissues, which largely avoids the unspecific side effects of systemic gene knockdown (33). Additionally, with the fusion of Cre recombinase to the ligand-binding domain of estrogen receptor (Cre-ER), the deletion of floxed genes can be induced at the desired time by tamoxifen administration (34). Taking advantage of these sophisticated tools, we constructed a mouse strain carrying the conditional LCMR1 allele with exons 2–4 flanked by loxP sites. We could achieve conditional gene inactivation of LCMR1 in different tissue or cell types by crossing the LCMR1flox mice with mice expressing specific Cre recombinase. In the present study, we bred LCMR1flox mice with Sftpc-CreERT2 mice harboring tamoxifen-inducible Cre activity specifically in AT2 cells. To our knowledge, this is the first reported AT2 cell-specific LCMR1 conditional knockout mouse model to date.
Surprisingly, all of the LCMR1ΔAT2 mice died within 17 days after tamoxifen administration in the absence of additional damaging agents, while all of the control mice were healthy and alive. We found massive AT2 cell loss in the lung tissues of LCMR1ΔAT2 mice through immunofluorescence staining for the AT2 cell marker, proSP-C. LCMR1 knockdown was reported to promote cell apoptosis in human osteosarcoma, laryngocarcinoma, and lung cancer cells in vitro (2-4). In addition, adipose-specific MED19 knockout in adult mice can cause lipodystrophy due to increased apoptosis and macrophage infiltration (12). Therefore, we hypothesized that AT2 cell loss in LCMR1ΔAT2 mice may be related to increased apoptosis. ProSP-C and TUNEL co-staining demonstrated more apoptotic AT2 cells in LCMR1ΔAT2 mice compared with those in LCMR1flox/flox mice, as evidenced simultaneously by proSP-C and cleaved caspase-3 double staining. Furthermore, the elevated apoptosis in AT2 cells was confirmed by upregulated p53 protein and downregulated Bcl-2 protein expression. The regulation of AT2 cell apoptosis has been reported in some studies but the main molecular mechanism remains uncertain (35,36). Therefore, further research is needed to determine the specific molecular mechanism through which LCMR1 deletion results in AT2 cell loss.
Here, we highlight two possible causes of death for LCMR1ΔAT2 mice. On the one hand, extensive AT2 cell loss resulting from LCMR1 deletion destroyed the integrity of the alveolar architecture, as evidenced by histopathological examinations. As essential components and stem cells of the alveolar epithelium, AT2 cells are important in alveolar maintenance and repair in the adult lung (14). Targeted injury of AT2 cells at a high level can cause lung injury, fibrosis, and even death in mice (37,38). On the other hand, the depletion of AT2 cells may also disrupt the homeostasis of pulmonary surfactant and weaken its function to lower the surface tension at the air-liquid interface. Despite the quantitative reduction of AT2 cells, we found dysmorphic or electron-dense lamellar bodies in AT2 cells through TEM, which may aggravate pulmonary surfactant dysfunction as lamellar bodies are functionally specialized organelles synthesizing and storing pulmonary surfactants in AT2 cells. SP-B and SP-C are the critical surfactant proteins involved in surfactant organization and its surface-active properties (39). We found significantly reduced expression of SP-B and SP-C in the lung tissues of LCMR1ΔAT2 mice compared with that of LCMR1flox/flox mice, especially SP-C, which was almost undetectable. Nevertheless, the expression of SP-D was significantly increased in the lung tissues of LCMR1ΔAT2 mice. One possible cause may be that AT2 cells are not the only source of SP-D which can also be synthesized and secreted by Clara cells and macrophages (40). Additionally, the changes in histopathology, permeability, and the inflammatory response observed in the lung tissues of LCMR1ΔAT2 mice produced a phenotype with features resembling lung injury. It was previously reported that SP-D expression was increased in the lung tissues of animals with acute lung injury (41,42).
Conclusions
In conclusion, this study demonstrated for the first time that the conditional knockout of LCMR1 specifically in the AT2 cells of adult mice results in a lethal phenotype with impaired lung structure and function. The main causes for this could lie in extensive AT2 cell loss, alveolar integrity destruction, and pulmonary surfactant deficiency. Our data indicate that LCMR1 is essential for AT2 cell integrity and maintenance. These findings also highlight the critical role of AT2 cells in maintaining lung homeostasis.
Acknowledgments
We thank the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Science for assistance with transmission electron microscopy.
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 30070335 and 30370616) and the China Postdoctoral Science Foundation (No. 20090461438).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-293/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-293/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-293/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-293/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments performed in this study were approved by the Institutional Animal Care Use Committee of Chinese PLA General Hospital (No. 2018-X14-25), in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen L, Liang Z, Tian Q, et al. Overexpression of LCMR1 is significantly associated with clinical stage in human NSCLC. J Exp Clin Cancer Res 2011;30:18. [Crossref] [PubMed]
- Xu Y, Li C, Tian Q, et al. Suppression of lung cancer metastasis-related protein 1 promotes apoptosis in lung cancer cells. Int J Mol Med 2012;30:1481-6. [Crossref] [PubMed]
- Yu W, Zhang Z, Min D, et al. Mediator of RNA polymerase II transcription subunit 19 promotes osteosarcoma growth and metastasis and associates with prognosis. Eur J Cancer 2014;50:1125-36. [Crossref] [PubMed]
- Zhao Y, Meng Q, Gao X, et al. Down-regulation of mediator complex subunit 19 (Med19) induces apoptosis in human laryngocarcinoma HEp2 cells in an Apaf-1-dependent pathway. Am J Transl Res 2017;9:755-61. [PubMed]
- Sato S, Tomomori-Sato C, Parmely TJ, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 2004;14:685-91. [Crossref] [PubMed]
- Boube M, Joulia L, Cribbs DL, et al. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 2002;110:143-51. [Crossref] [PubMed]
- Richter WF, Nayak S, Iwasa J, et al. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat Rev Mol Cell Biol 2022;23:732-49. [Crossref] [PubMed]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 2010;11:761-72. [Crossref] [PubMed]
- Soutourina J. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 2018;19:262-74. [Crossref] [PubMed]
- Dean JM, He A, Tan M, et al. MED19 Regulates Adipogenesis and Maintenance of White Adipose Tissue Mass by Mediating PPARγ-Dependent Gene Expression. Cell Rep 2020;33:108228. [Crossref] [PubMed]
- Immarigeon C, Bernat-Fabre S, Guillou E, et al. Mediator complex subunit Med19 binds directly GATA transcription factors and is required with Med1 for GATA-driven gene regulation in vivo. J Biol Chem 2020;295:13617-29. [Crossref] [PubMed]
- Ding N, Tomomori-Sato C, Sato S, et al. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem 2009;284:2648-56. [Crossref] [PubMed]
- Rock JR, Barkauskas CE, Cronce MJ, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A 2011;108:E1475-83. [Crossref] [PubMed]
- Barkauskas CE, Cronce MJ, Rackley CR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 2013;123:3025-36. [Crossref] [PubMed]
- Chen Q, Liu Y. Heterogeneous groups of alveolar type II cells in lung homeostasis and repair. Am J Physiol Cell Physiol 2020;319:C991-C996. [Crossref] [PubMed]
- Basil MC, Katzen J, Engler AE, et al. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell 2020;26:482-502. [Crossref] [PubMed]
- D’Agnillo F, Walters KA, Xiao Y, et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci Transl Med 2021;13:eabj7790. [Crossref] [PubMed]
- Lechner AJ, Driver IH, Lee J, et al. Recruited Monocytes and Type 2 Immunity Promote Lung Regeneration following Pneumonectomy. Cell Stem Cell 2017;21:120-134.e7. [Crossref] [PubMed]
- Li FJ, Surolia R, Li H, et al. Citrullinated vimentin mediates development and progression of lung fibrosis. Sci Transl Med 2021;13:eaba2927. [Crossref] [PubMed]
- Gerard L, Lecocq M, Bouzin C, et al. Increased Angiotensin-Converting Enzyme 2 and Loss of Alveolar Type II Cells in COVID-19-related Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med 2021;204:1024-34. [Crossref] [PubMed]
- Agaësse G, Barbollat-Boutrand L, Sulpice E, et al. A large-scale RNAi screen identifies LCMR1 as a critical regulator of Tspan8-mediated melanoma invasion. Oncogene 2017;36:446-57. [Crossref] [PubMed]
- Zhang X, Gao D, Fang K, et al. Med19 is targeted by miR-101-3p/miR-422a and promotes breast cancer progression by regulating the EGFR/MEK/ERK signaling pathway. Cancer Lett 2019;444:105-15. [Crossref] [PubMed]
- Wang K, Wang X, Fu X, et al. Lung cancer metastasis-related protein 1 promotes the transferring from advanced metastatic prostate cancer to castration-resistant prostate cancer by activating the glucocorticoid receptor α signal pathway. Bioengineered 2022;13:5373-85. [Crossref] [PubMed]
- Zhang Y, Qin P, Xu X, et al. Mediator Complex Subunit 19 Promotes the Development of Hepatocellular Carcinoma by Regulating the AKT/mTOR Signaling Pathway. Front Oncol 2021;11:792285. [Crossref] [PubMed]
- Zhang X, Wu J, Hu C, et al. CXCL11 negatively regulated by MED19 favours antitumour immune infiltration in breast cancer. Cytokine 2022;162:156106. [Crossref] [PubMed]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823-6. [Crossref] [PubMed]
- Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science 2018;361:866-9. [Crossref] [PubMed]
- Bai Y, Liu Y, Su Z, et al. Gene editing as a promising approach for respiratory diseases. J Med Genet 2018;55:143-149. [Crossref] [PubMed]
- Moses C, Kaur P. Applications of CRISPR systems in respiratory health: Entering a new 'red pen' era in genome editing. Respirology 2019;24:628-637. [Crossref] [PubMed]
- Sreedurgalakshmi K, Srikar R, Rajkumari R. CRISPR-Cas deployment in non-small cell lung cancer for target screening, validations, and discoveries. Cancer Gene Ther 2021;28:566-580. [Crossref] [PubMed]
- Liu X, Zhao X, Yuan Y, et al. Accurate detection of lung cancer-related microRNA through CRISPR/Cas9-assisted garland rolling circle amplification. J Thorac Dis 2022;14:4427-4434. [Crossref] [PubMed]
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A 1992;89:6861-5. [Crossref] [PubMed]
- Liu B, Jing Z, Zhang X, et al. Large-scale multiplexed mosaic CRISPR perturbation in the whole organism. Cell 2022;185:3008-3024.e16. [Crossref] [PubMed]
- Feil R, Brocard J, Mascrez B, et al. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A 1996;93:10887-90. [Crossref] [PubMed]
- Olajuyin AM, Zhang X, Ji HL. Alveolar type 2 progenitor cells for lung injury repair. Cell Death Discov 2019;5:63. [Crossref] [PubMed]
- Zheng J, Zhu S, Xu H, et al. miR-363-3p inhibits rat lung alveolar type II cell proliferation by downregulating STRA6 expression and induces cell apoptosis via cellular oxidative stress and G1-phase cell cycle arrest. Transl Pediatr 2021;10:2095-2105. [Crossref] [PubMed]
- Garcia O, Hiatt MJ, Lundin A, et al. Targeted Type 2 Alveolar Cell Depletion. A Dynamic Functional Model for Lung Injury Repair. Am J Respir Cell Mol Biol 2016;54:319-30. [Crossref] [PubMed]
- Sisson TH, Mendez M, Choi K, et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am J Respir Crit Care Med 2010;181:254-63. [Crossref] [PubMed]
- Milad N, Morissette MC. Revisiting the role of pulmonary surfactant in chronic inflammatory lung diseases and environmental exposure. Eur Respir Rev 2021;30:210077. [Crossref] [PubMed]
- Voorhout WF, Veenendaal T, Kuroki Y, et al. Immunocytochemical localization of surfactant protein D (SP-D) in type II cells, Clara cells, and alveolar macrophages of rat lung. J Histochem Cytochem 1992;40:1589-97. [Crossref] [PubMed]
- Sakamoto K, Hashimoto N, Kondoh Y, et al. Differential modulation of surfactant protein D under acute and persistent hypoxia in acute lung injury. Am J Physiol Lung Cell Mol Physiol 2012;303:L43-53. [Crossref] [PubMed]
- Yan XX, Zheng AD, Zhang ZE, et al. Protective effect of pantoprazole against sepsis-induced acute lung and kidney injury in rats. Am J Transl Res 2019;11:5197-211. [PubMed]
(English Language Editor: J. Gray)


