ADCY9 functions as a novel cancer suppressor gene in lung adenocarcinoma
Highlight box
Key findings
• ADCY9 gene may serve as a novel target in suppressing lung adenocarcinoma.
What is known and what is new?
• Partial function of ADCY9 gene is known.
• The correlation of ADCY9 gene and lung adenocarcinoma was added in this research.
What is the implication, and what should change now?
• The ADCY9 gene may be a potential biomarker in predicting lung adenocarcinoma or may provide a new thinking for treating lung adenocarcinoma
Introduction
Based on Global Cancer Statistics 2020, lung cancer still had the second highest diagnosed incidence among all cancer types and still has the highest cancer-related mortality rate all over the world, with an estimated 2 million new cases and 1.8 million deaths per year (1,2). In multiple pathological types of lung cancer cases, non-small cell lung cancer (NSCLC), particularly lung adenocarcinoma (LUAD), accounts for the largest proportion of pathological types, resulting in nearly 1.35 million deaths worldwide each year (3). To improve the accuracy of early diagnosis and treatment efficacy in LUAD, high-throughput sequencing, enhanced CT scans, and biomarker molecular targeted therapy are widely used (4). It was reported that TP53 missense but not-nonsense mutants are sensitive to immune checkpoint inhibitors therapy like nivolumab (antiPD-L1) plus ipilimumab (antiCTLA-4) (5). Novel targeted agents, such as the covalent KRAS G12C inhibitors, may allow a tailored treatment for LUAD patients with KRAS mutations (6). Further, evidence from the public database showed that distinct combinations of STK11, EGFR, and TP53 mutations were major determinants of the tumor immune profile (TIP) and highly correlated with the immune therapy consequences (7). Reports also showed that LaminB1, a member of the intermediate filaments supergene family, could be a biomarker for a worse prognosis in LUAD (8). Further, CTSL2, a lysosomal cysteine protease, is considered to be a biomarker of tumor metastasis in LUAD patients (9). Even though great progress has been made in developing new molecular biomarkers and therapeutic targets, only 15.6% of all patients with lung cancer are alive 5 years or more after the diagnosis (10). Hence, identifying a new biomarker is of great significance to guide clinicians in improving the diagnosis and prognosis of LUAD.
ADCY9 is a member of the cyclase gene family located on chromosome 16p13.3 (11). ADCY9 is primarily expressed in lung, thyroid (12), large-B cell (13), and 24 kinds of other tissues. As a “membrane-bound” enzyme, activation of adenylyl cyclase type 9 hydrolyzes ATP, leading to the intracellular production of cyclic AMP (cAMP, adenosine-3', 5'-monophosphate), which plays a critical role in regulating GPCR signal pathway transduction as a second messenger (14). Research validated that the ADCY9 gene was identified as a potential gene related to the development of certain diseases. For instance, stroke (15) and malaria (16) were reported to be concerned with ADCY9 genetic polymorphisms. Loss of the ADCY9 gene is associated with the imbalance in the immune regulatory response, leading to immune-mediated diseases, such as allergic disorders and asthma (17). Silence of ADCY9 contributes to the reduction in aortic atherosclerosis in the absence of CETP (cholesteryl ester transfer protein) (18,19). However, research on the ADCY9 gene in lung cancer is not sufficient. Therefore, we aimed to examine the relationship between ADCY9, expression level, and clinicopathological features in LUAD via Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) lung cancer database, and cellular function assays.
In this research, to explore the specific function of ADCY9 expression, we analyzed the prognostic value in LUAD patients. We then estimated its expression level in cell lines and detected its biological functions by altering its expression in vitro through SPCA1 and A549 cell lines. Our findings may be helpful in predicting the prognosis of patients with LUAD and in directing clinical chemotherapy and immunotherapy in the future, which may contribute to early diagnosis and increase the overall survival (OS) of patients with LUAD. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1027/rc).
Methods
Acquisition of data sources and preconditioning
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The gene expression quantification profile, miRNA expression profile, and clinical files of LUAD were obtained from GEO and TCGA dataset. ADCY9 gene was selected via the survival analysis filter performed on the GEO database GSE42127 containing 154 LUAD tissues and 23 normal lung tissues and further validated through data acquired from TCGA database containing 497 LUAD tissues and 54 normal lung tissues. ADCY9-related microRNA-targeted LncRNA was downloaded from the ENCORI database (Encyclopedia of RNA Interactomes). Samples that lacked clinical data and survival data less than or equal to zero were removed from this research.
Clinical features and prognostic analysis of ADCY9 in LUAD
The survival analysis curve was drafted from data acquired from GEO and TCGA databases using the Kaplan-Meier (K-M) method to compare the survival time with the ADCY9 expression level of LUAD patients, along with “survival” and “survminer” packages in R. ADCY9 expression level in different clinical feature groups like age, gender, and stage was realized using the “ggpubr” package in R software. Receiver operating characteristic (ROC) curve analysis is a comprehensive index reflecting the continuous variables of sensitivity and specificity; the larger the area under the curve, the better the diagnostic accuracy. R software packages ‘survival’, ‘survminer’, and ‘timeROC’ were used to realize the ROC image (20).
Co-expression analysis of ADCY9 and its related miRNA and lncRNA
To further explore the downstream mechanism of ADCY9, the Wilcox test and the R software packages “limma”, “ggpubr”, “ggextra”, and “reshape2” were adopted to find ADCY9 combining miRNAs based on the threshold of corFilter =0.2 and P value filter =0.01. The miRNA-lncRNA and lncRNA-ADCY9 targeting relationship was realized through the same packages above based on the threshold of corFilter =0.2 and P value filter =0.001. The correlation between the immune cell biomarkers level and ADCY9 was calculated by R software packages “limma”, “ggpubr”, “ggextra”, and “reshaped”. The relationship between ADCY9 expression level and immune cells was evaluated using the TIMER2.0 database online web tools, which were used to analyze the variance in the copy number of the ADCY9 gene and the immune cells recruiting level.
Functional enrichment analysis
Gene Ontology (GO), including biological process, molecular function, and cell composition combined with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, was to explore the distribution of genes in specific pathways (21). BiocManager packages ‘org.Hs.eg.db’, ‘DOSE’, ‘clusterProfiler’, and ‘enrichplot’ were used to carry out GO and KEGG analyses, followed by the step of visualization through ‘colorspace’, ‘stringi’, and ‘ggplot2’ packages in R software. To research the potential biological pathways among ADCY9 low-expressed and high-expressed patients in the progression of LUAD, multiple gene enrichment analysis was carried out through gene set enrichment analysis (GSEA) coupled with the ggplot package in R (22).
Univariate and multivariate Cox analysis
According to the data from GEO and TCGA databases, univariate and multivariate Cox regression analysis was conducted to explore which was the most relevant clinical factor in the survival of LUAD patients through the “survival” package in R (23,24). Age, gender, stage, T, N, M classification, and ADCY9 expression were also contained in this model.
Pan-cancer analysis
Pan-cancer analysis, including alteration frequency and expression level in other cancer types along with protein expression level of ADCY9 in LUAD and OS of ADCY9 in LUAD, were analyzed using the web tools of TIMER and the CPTAC database.
Cell culture and in vitro overexpression
The human LUAD cell lines A549, HCC827, and H23 were purchased from the Shanghai Cell Bank of the Chinese Academy of Medical Sciences (CAMS). Normal human pulmonary epithelial cell lines BEAS-2B and H1299 were purchased from the American Type Culture Collection (ATCC, USA). SPCA1 was purchased from Nanjing Keygen Biotech Co., Ltd. (KG092, China). Reports suggested that SPCA1 cell lines were contaminated by the Hela cell line. However, the SPCA1 cell line supplied by Keygen Biotech was provided with an STR cell identification report that validated that their cell line was free from cross-contamination with other human cell lines. All the cell lines were cultured in the medium, which consisted of RPMI-1640 (11875093, Gibco, Thermo Fisher, USA), 10% fetal bovine serum (10099141C, Gibco), 0.1 mg/mL streptomycin (10378016, Gibco) and 100 U/mL penicillin (10378016, Gibco), and was maintained under 37 ℃ at 5% CO2 atmosphere.
The ADCY9 coding sequence was cloned into pcDNA3.1 (RIBOBIO, China) to construct the ADCY9 overexpression vector, and the empty vector was used as a negative control. Lipofectamine3000 Transfection Reagent was purchased from (L3000008, Thermo Fisher). The Lipofectamine3000 kit (L300000, Thermo Fisher) was used to perform transfection in accordance with the operating instructions in different LUAD cell lines. The transient transfection efficiency was tested 48 h later via quantitative real-time polymerase chain reaction (qRT-PCR). The SPCA1 and A549 cell lines were selected to carry on stable transfection. The stably transfected cell lines (SPCA1 and A549) were filtered by G418 (1:10,000) and the efficiency of stable transfection was evaluated using qRT-PCR and western blot.
RNA extraction and qRT-PCR
Both cell lines and tissues (80 pairs) were treated with Trizol reagent (10296028, Thermo). The process of reverse transcription into cDNA was done using the method of the RT Reagent kit (18091200, Thermo) according to the manufacturer’s protocol. The mRNA expression level was estimated using a ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., China). GADPH was taken as an internal reference. Primers adopted are listed below: ADCY-9 forward: 5'-TAAGATGCAGCAGATCGAAGAAGTCAG-3'; ADCY-9 reverse: 5'-AGATCGTTCAGGAGACCCACCAG-3'; GAPDH forward: 5'-GTCTCCTCTGACTTCAACAGCG-3' GAPDH reverse: 5'-ACCACCCTGTTGCTGTAGCCAA-3'.
The procedures of qRT-PCR were 95 ℃ for 5 min, and the subsequent step is 40 cycles at 95 ℃ for 15 seconds, 60 ℃ for 30 seconds, and 72 ℃ for 30 seconds. The assay was tested 3 times, and the comparative quantification cycle (Cq) method (2−ΔΔCq) was used to estimate the relative expression level of the ADCY9 gene.
Western blot assays
RIPA lysis buffer (P0013C, Beyotime, China) containing 1% phosphatase and protease inhibitors (PMSF) was used to lyse all the cell lines and the overexpressed SPCA1 and A549 cell lines. We applied 10% sodium dodecyl sulfate-polyacrylamide for the lower gel and 4% sodium dodecyl sulfate-polyacrylamide for the upper gel to separate the protein samples electrophoresis then transferred this to the PVDF membrane (Whatman; Sigma-Aldrich, USA) under the current of 300 mA for 150 min. We used 5% skimmed milk to block the nitrocellulose membranes for 2 h at room temperature, followed by the step of specific antibody incubation overnight at 4 ℃. The specific antibodies immunoblotting with the PVDF membrane were anti-ADCY9 (1:1,000, AP50059, Abcepta, USA) and anti-GAPDH (1:10,000, 10494-1-AP, Proteintech, USA). Afterward, the PVDF membranes were washed in the Tris-buffered saline buffer (1XTBST) 3 times for 10 min and treated with 1:10,000 horseradish peroxidase (HRP) conjugated secondary antibody (1:1,000, SA00004-2, Proteintech) for 2 h at room temperature. The subsequent step was washing the PVDF membrane 3 times for 5 min, and an ECL (Electro-Chemi-Luminescence) kit mixture at the proportion of 1:1 was used to visualize the bands on the odyssey imaging system. The relative expression quantity of proteins was the specific value of the target protein relative to the internal reference protein. The experiment was carried out 3 times to guarantee the reproducibility of the result.
Immunochemistry
Formalin was used to treat tumor tissue sections (4 µm thick), followed by bedding the sections in paraffin. Xylene anhydrous ethanol was used to remove paraffin while catalase was blocked by 3% hydrogen peroxide. The sections were incubated with ADCY9 antibodies (1:1,000, ab191423, Abcam, UK) for 24 h at 4 ℃, then sections were incubated with secondary antibodies (1:1,000, SA00004-2, Proteintech) for 2 h at 37 ℃. The nuclei were stained with hematoxylin (H8070, Solarbio, China) after DAB kit (K5007, DAKO, Denmark) staining. Image J was used to calculate the integrated density of the DAB staining positive area to estimate the expression of ADCY9.
Flow cytometry assays of cell cycle distribution
The cellular cycle experiment was performed according to the protocol of the manufacturer’s DNA Content Quantitation Assay (Cell Cycle, CA1510, Solarbio). We washed the SPCA1 and A549 vector and SPCA1, A549 overexpressed cell lines with PBS once, collected it and centrifuged it at 1,500 rpm for 5 min, then used a cell counter to adjust the cell concentration to 1×106/mL, and took 1 mL of single-cell line suspension. After the single-cell suspension was centrifuged, we removed the supernatant, added 500 µL of 70% pre-cooled ethanol to the cells, and fixed it for 2 h to overnight, stored at 4 ℃, before washing off the fixative with phosphate buffered saline (PBS) before staining. If necessary, the cell suspension was filtered once with a cell sieve. A total of 100 µL of RNaseA solution was added to the cell precipitation, which resuspended the cells. Then, this solution was bathed in water at 37 ℃ for 30 min. We added 400 µL of propidium iodide (PI) staining solution and mixed well, incubated the solution at 4 ℃ for 30 min in the dark, tested the flow cytometer, and recorded the red fluorescence at the excitation wavelength of 488 nm. The experiment was carried out 3 times to ensure the reproducibility of the result.
Flow cytometry of cellular apoptosis
Cellular apoptosis assay was performed in accordance with the protocol of the manufacturer’s Annexin V-FITC Apoptosis Detection Kit (Cell Apoptosis, CA1020-50T, Solarbio). The SPCA1 and A549 vector and SPCA1, A549 overexpression cell lines were collected and then centrifuged at approximately 1,000 rpm for 5 min to precipitate the cells. After removing the supernatant, we added 50 µL of culture medium to avoid aspiration of cells. About 1 mL of 4 ℃ pre-cooled PBS was added, resuspending the cells, then the solution was centrifuged again to precipitate the cells and remove the supernatant. The binding buffer was diluted to a 1:9 ratio with deionized water (2 mL 10× binding buffer + 18 mL deionized water), then the cells were resuspended with 1× binding buffer, and the concentration was adjusted to 1–5×106/mL. Before adding 5 µL Annexin V/FITC, 100 µL cell suspension was taken into a 5 mL flow tube, followed by the step of mixing well and incubating at room temperature for 5 min in the dark, adding 5 µL PI solution and 400 µL PBS, performed on the flow cytometry or fluorescence microscopy immediately and performed 3 times to makes sure the repeatability of an experimental outcome.
Measurements of cell proliferation assays
The SPCA1, A549 and SPCA1, A549 overexpression cell lines were digested by trypsin (25200072, Thermo Fisher), followed by suspending the cell lines. Both the SPCA1, A549 cell line and the SPCA1, A549 overexpression cell lines were seeded in 96-well cell plates at the density of 2000 each well and separately cultivated for 30 wells, with 6 wells a line and 5 rows in total. After culturing for 24 h, each row was treated with 10 µL CCK-8 reagent and maintained at 37 ℃ for another 2 h. Under the absorption light of 450 nm, the optical density (OD) value was recorded by the microplate reader. The same procedures were repeated for 6 days. We repeated the same procedure 3 times.
Monoclonal formation assays
Trypsin (25200072, Thermo Fisher) was used to digest the SPCA1, A549 and SPCA1, A549 overexpression cells to prepare for the resuspension. After calculated by the cell counter, at the density of 500 cells per well, cell lines were cultivated into the six-well plated and each well was developed by 1.5 mL medium which is made up of 10% bovine serum and 90% RPMI1640 (11875093, Gibco, Thermo Fisher) at 37 ℃ with 5% CO2 for 2 weeks. The process was not terminated until the colony of cell lines could be visualized. The next step was immobilizing the cells with 4% paraformaldehyde for 24 h, followed by staining with 0.1% crystal violet for 30 min. The assay was repeated 3 times before calculating the number of cell groups by ImageJ.
Transwell chamber assays of cell migration and invasion
The SPCA1, A549 and SPCA1, A549 overexpression cell lines were harvested via trypsin (25200072, Thermo Fisher) and resuspended. Cells were added into the upper chamber at the density of 5×105 per chamber. Thereafter, the medium RPMI1640 was added into the upper chamber at the volume of 500 µL, while 20% bovine serum medium was added into the lower chamber at the same volume. Both cell lines migrated or invaded for 48 h. After transfection for 48 h, the remaining cells in the upper chamber were wiped off by cotton swabs. Thereafter, the cells on the bottom chambers were immobilized with 4% paraformaldehyde for 2 h and stained with 0.4% crystal violet for 30 min. The assay was repeated 3 times, and the number of the transfected cells was visually observed in 9 random areas under the bright field of a microscope and estimated via image J.
Statistical analysis
Two group comparisons were carried out via Welch’s t-test. The K-M survival curve was generated by median gene separation or a combination of gene expression (patients with ADCY9 and ADCY9 expression in the top 50% and ADCY9 and ADCY9 expression in the lowest 50%) and OS time was used to generate a K-M survival curve for survival comparison. Correlation assessment was generated by Pearson’s correlation test and the Wilcox test. P values <0.05 were considered statistically significant.
Results
Correlation of ADCY9 expression level and prognosis in LUAD patients
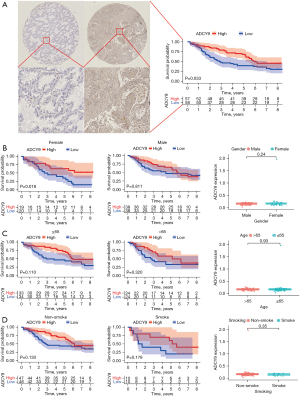
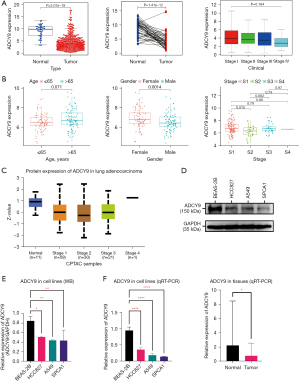
To explore whether the expression of ADCY9 is linked with the OS of LUAD patients, first, we used bioinformatics approaches to analyze the survival of LUAD patients from the GEO and TCGA dataset. The median ADCY9 expression level was adopted as the value to classify the patients into the ADCY9 high expression group and the ADCY9 low expression group. Then, we conducted the K-M method to compare the OS of the 2 groups. As shown in Figure 1A (P<0.001) and Figure 1B (P=0.017), among the 2 groups, the OS was significantly higher in the group in which ADCY9 was more highly expressed and the pan-cancer analysis of ADCY9 expression was displayed in Figure 1C (LUAD = 515; Normal = 59). The survival curve of ADCY9 in other types of cancers such as renal clear cell carcinoma (RCCC), lung squamous cell carcinoma (LUSC), bladder cancer (BCA) and brain lower grade glioma (LGG) were exhibited in Figure 1D and alteration of frequency was also shown in Figure 1E. Judging from the results of the LUAD tissue microarray (LUAD specimens n=115), higher expression of ADCY9 may indicate a better prognosis of LUAD patients (P=0.033; Figure 2A). However, the OS is statistically significant with expression level of ADCY9 in female group while the male group turns the opposite (Figure 2B). Meanwhile, OS is not statistically significant with expression level of ADCY9 in terms of age and smoking history (Figure 2C,2D). Meanwhile, ADCY9’s prognostic value needs to be further explored in LUAD. Therefore, univariate and multivariate Cox proportional hazards regression were performed to analyze the clinical data of LUAD patients from the GEO and TCGA database (Figure 3). According to the univariate Cox proportional hazards regression, ADCY9 (P<0.01), stage (P<0.01), and TMN clinical stage (P<0.05) were identified as prognostic factors for OS by univariate analysis. Multivariate analysis was also carried out to identify that ADCY9 (P=0.005; Figure 3B; P<0.001; Figure 3C) and clinical stage (P=0.006, Figure 3B; P=0.028, Figure 3C) may be deemed as independent prognostic factors for OS in patients with LUAD. The ROC curve demonstrates that the ADCY9 may be a precise factor to predict the prognosis of LUAD (Figure 3D).


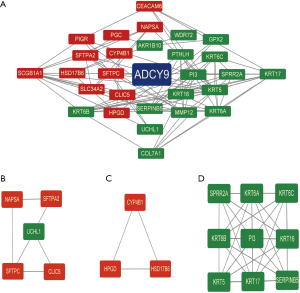
Bioinformatics analysis of ADCY9-related genes, miRNAs and miRNA-related lncRNAs
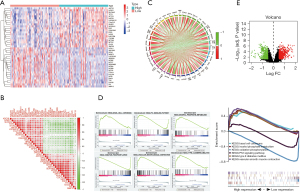
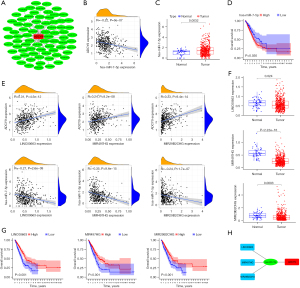
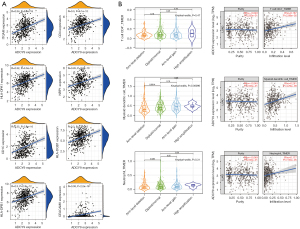
Functional enrichment reveals that ADCY9 and its related genes were primarily enriched mitotic nuclear division and organelle fission pathway (Figure 3A). Correlation analysis, circus plot, heatmap diagram and volcano diagram exhibits the relevance of ADCY9 and its related genes while GSEA reveals the pathway that ADCY9 gene majorly enriched in and protein-protein interaction (PPI) network (Figure 4). PPI network and regarding mcodes were established based on ADCY9 and its related genes (Figure 5). Correlation analysis was used to perform PPI network of the ADCY9 and its related miRNAs and miRNA-related lncRNAs, as shown in Figure 6A, among which miRNA hsa-miR-7-5p was validated to have a negative correlation with the expression of ADCY9 (Figure 6B) and showed a significant distinction between normal and tumor tissues (Figure 6C), meanwhile, exhibiting a lower survival rate in miRNA hsa-miR-7-5p high expression groups (Figure 6D). High expression of hsa-miR-7-5p-related lncRNAs are positively correlated with the expression of ADCY9 while negatively correlated with the expression of miRNA hsa-miR-7-5p (Figure 6E). As shown in the Figure 6F, lncRNAs LINC00663 (P<0.05), MIR29B2CHG (P<0.001), and MIR497HG (P<0.0001) had obvious differences between normal and tumor tissues and may have a positive effect on the OS of the LUAD patients (P<0.001; Figure 6G). The CeRNA mechanism PPI network between miRNA hsa-miR-7-5p and its relevant lncRNAs (Figure 6H) were analyzed using the bioinformatics method.
Correlation analysis between ADCY9 gene and relevant immune cells infiltrating
Links between the ADCY9 gene and immune cell biomarkers were explored through TCGA dataset using R software. The results showed that high expression of ADCY9 may have a positive effect on the recruiting of CD4+ T cells, neutrophils, and dendritic cells because markers CEACAM8/ITGAM and HLA-DPB1/HLA-DQB1/HLA-DRA/ HLA-DPA1/CD1C/NRP1/ITGAX are specific biomarkers of neutrophils and dendritic cells (Figure 7A). The TIMER2.0 database was used to verify the outcomes above and acquired the same result (Figure 7B). The copy number of the ADCY9 gene was analyzed through the TIMER2.0 database demonstrated that arm-level deletion of the ADCY9 gene significantly decreased in recruiting dendritic cells compared with the diploid/normal group, while an obvious decrease was not discovered in recruiting CD4+ T cells and neutrophils between arm-level deletion group and diploid/normal group (Figures 7B).
ADCY9 expression level in tumor and normal tissues
The expression level was first analyzed via data obtained from TCGA database. As shown in Figure 8A, the expression of ADCY9 in normal tissues (n=54) exhibited significantly higher levels than that in LUAD tissues (n=497; P<0.001) while expression of ADCY9 is not statistically significant among different stages of LUAD (Figures 8A). According to the data from GEO database, expression of ADCY9 is statistically significant among different gender groups while shows opposite conclusion in terms of age and stage (Figure 8B). Further, protein expression level of ADCY9 from CPTAC (Figure 8C; LUAD = 111; Normal = 11) had the same results, which showed that ADCY9 was more highly expressed in normal tissues than tumor tissues (P<0.001). In addition to bioinformatic analysis, western blot (P<0.01; Figure 8D,8E) showed a higher protein expression level of ADCY9 in the normal cell lines, and qRT-PCR, showed a higher mRNA expression level of ADCY9 both in normal cell lines (P<0.001; Figure 8F) and in normal tissues (P<0.05; Figure 8F). These outcomes suggest that the ADCY9 gene is significantly more highly expressed in normal tissues than tumor tissues, and up-regulation of the ADCY9 gene may be associated with a better prognosis of LUAD.
The expression level of the ADCY9 gene is associated with the proliferation and migration of LUAD
The over-expression level of ADCY9 in both SPCA1 and A549 cell line was validated by qRT-PCR and western blot assays (Figure 9A,9B). To explore the potential effect of ADCY9 on LUAD cell proliferative abilities, CCK-8 and clone formation assays were performed. Outcomes of CCK8 indicated that the OD values of the ADCY9 overexpression group were meaningfully lower than the vector group in SPCA1, A549 cell lines from 0 to 120 h (Figure 9C,9D). From the clone formation assay, we observed that, in comparison with the vector group in SPCA1, A549 cell lines, the ADCY9 overexpression group showed a visible decrease in colony number (P<0.05; Figure 9E,9F), which suggests that ADCY9 overexpression may repress proliferation of LUAD.
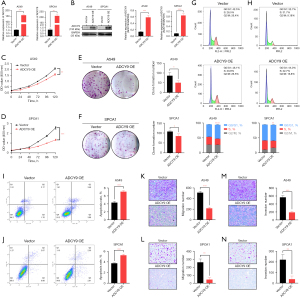
On the basis of data from flow cytometry, over-expression of ADCY9 inhibited the cell cycle because the ADCY9 over-expression group possessed a higher percentage in the Synthetic period (P<0.0001; Figure 9G; P<0.01; Figure 9H). Further, compared to the vector group, the ADCY9 overexpression group promoted cell apoptosis according to Figure 9I (P<0.001) and Figure 9J (P<0.001). As shown in migration and invasion assays, compared to the SPCA1, A549 vector group, the ADCY9 overexpression group displayed nearly a 2-fold decrease in terms of migration and invasive cell number (P<0.001 Figure 9K; P<0.01; Figure 9L; P<0.001; Figure 9M; P<0.01; Figure 9N). This evidence illustrates that ADCY9 may be a biomarker that suppresses the proliferation, migration, and metastasis of LUAD.
Discussion
LUAD is still the leading cause of death among all cancer types (25). It accounts for almost 25% of all cancer mortalities (26). Moreover, as the major pathological subtype of lung cancer, LUAD occurs in 39% of newly diagnosed lung cancer cases, leading to 1.35 million deaths worldwide each year (2,3). In the past decade, advanced treatments such as PD-1 molecular therapy and immunotherapy, to certain degrees, have improved the prognosis of LUAD patients (27-29). Yet, the relative survival rate of lung cancer is far from ideal, and merely 15.6% of all patients with lung cancer can live 5 years or more after their diagnosis (10). The mechanisms of tumorigenesis and metastasis still remain ambiguous (30-32). Society and patients have undertaken huge expenses to treat LUAD; therefore, we are hoping to offer a new biomarker to develop targeted therapies and diagnosis methods for LUAD in the future.
The only membrane-bound AC subtype that is not sensitive to adenylate cyclase activator (AC), mainly composed of 9 isoforms (33,34), is an integral membrane protein that can convert ATP into cAMP, causing cell signal responses, and is an effector in the G protein coupling system, which is widely distributed in the cell membrane of mammals (35-37). Among all the subtypes of adenylate cyclase, ADCY9 is the only membrane-bound AC subtype that is not sensitive to the adenylate cyclase activator. It is capable of being stimulated by beta-adrenergic receptor activation and is insensitive to forskolin, calcium, and somatostatin, which contributes to the CRH, corticosteroids and beta-adrenergic signaling pathway cascades (34,38,39). Research shows that adenylyl cyclase 9 is linked to HDL function and may result in cardiovascular events in multiple pathologies (39,40). Downregulation of ADCY9 is related to positive effects on atherosclerosis and cardiovascular endothelial function and a gain in adipose tissue volume in the absence of CETP (18,41). Apart from cardiovascular disease, the ADCY9 gene is linked to Rubinstein-Taybi syndrome, a rare congenital, plural formative, and neurodevelopmental disorder (42). Previous research into ADCY9 also validated that its expression level is associated with poor prognosis in colorectal cancer, hepatocellular carcinoma, and glioma occurrence (43-45). Meanwhile, specific studies were carried out to discover the correlation between the ADCY9 gene and LUAD. It was reported that ADCY9 might be associated with the estrogen signaling pathway in LUAD, which may improve the prognosis of LUAD patients (46). Further, research exhibited that the Phospholipase C Signaling network, including mutated genes like ADCY9, ADCY2, and GNB5, may indicate the facilitation of lung metastasis from colon cancer (47). In recent years, the competing endogenous RNAs (ceRNAs) mechanism has become a research highlight in terms of tumor progression regulating. With the in-depth study of genomics, the regulatory relationship between miRNA and target genes and the ceRNA mechanism between miRNA, lncRNA and mRNA have become a hot topic in the research of tumor progression (48), especially in lung cancer, lncRNA JPX competitively sponging miRNA miR-33a-5p has been validated to be related with epithelial-mesenchymal transition (EMT) in lung cancer. However, research on the ADCY9 gene and its related mechanism of specific miRNA and lncRNA have rarely been performed. It was reported that ZEB1 relieved strong basal repression of α1 integrin (ITGA1) mRNA, which in turn upregulated ADCY9 by sponging miR181b (49). In addition, reports also show that coupled with NMUR1 and SYT1genes, the ADCY9 can be a target gene of 2 miRNAs (hsa-miR-490-3p and hsa-miR-1293) and participates in 2 circRNA–miRNA interactions including 2 circRNAs (hsa_circ_0008234 and hsa_circ_0002360) and 2 miRNAs (hsa-miR-490-3p and hsa-miR-1293), which may be involved in pathogenesis and prognosis in patients with LUAD (50). Judging from research concerning ADCY9 and its related miRNA or lncRNA, the ADCY9 gene may have the potential to be the biomarker or therapeutic target in LUAD. The bioinformatics analysis combined with data mining appears to be an approach to discovering potential biomarkers in cancer research (51,52). Currently, new bioinformatics analyses focus on the correlation between gene expression patterns, representative PPIs, and the potential for clinical metastases to uncover novel survival-related sub network signatures as a function of tumor proliferative potential (53). Single-cell Multi-omics Gene co-Regulatory algorithm (SMGR) is used to detect coherent functional regulatory signals and target genes from the joint single-cell RNA-sequencing (scRNA-seq), and single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) data obtained from different samples is used to explore gene expression and tumor progression (54). In this research, via data mining from TCGA and the GEO database, we observed that the expression level of ADCY9 was significantly higher in adjacent normal tissue than in LUAD tissue. The analysis also explored the association between the ADCY9 expression level with age, gender, and T classification. The result of the GSEA analysis suggested that overexpression of ADCY9 might promote basal cell carcinoma and phosphate metabolism while inhibiting oxidative phosphorylation. Validation was conducted through western blot, qRT-PCR, and cellular function experiments, the results of which exhibited both significant diagnostic and prognostic value in LUAD. Although the ADCY9-miRNA, miRNA-lncRNA, and ADCY9-lncRNA targeting relationship was analyzed using bioinformatics methods, further experiments are required to verify the gene-miRNA targeting relationship and the ceRNA mechanism.
Conclusions
The present research illustrates that ADCY9 is highly expressed in adjacent normal tissue and is linked with a better survival rate in LUAD. Moreover, this is the first time a report validated that the overexpression of the ADCY9 gene is capable of affecting the invasion and proliferation of LUAD, indicating that ADCY9 may function as a novel cancer suppressor gene in the diagnosis and therapy of LUAD. These findings indicate that ADCY9 might be a potential biomarker for LUAD treatment and can provide a new approach for targeted therapy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81770266), “Six-one” Project for High-level Health Talents (No. LGY2016037), Nantong Key Laboratory of Translational Medicine in Cardiothoracic Diseases, Nantong Clinical Medical Research Center of Cardiothoracic Disease, and the Institution of Translational Medicine in Cardiothoracic Diseases in Affiliated Hospital of Nantong University.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1027/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1027/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1027/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Sun H, Liu SY, Zhou JY, et al. Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine 2020;60:102990. [Crossref] [PubMed]
- Ghimessy A, Radeczky P, Laszlo V, et al. Current therapy of KRAS-mutant lung cancer. Cancer Metastasis Rev 2020;39:1159-77. [Crossref] [PubMed]
- Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin Cancer Res 2018;24:5710-23. [Crossref] [PubMed]
- Li W, Li X, Li X, et al. Lamin B1 Overexpresses in Lung Adenocarcinoma and Promotes Proliferation in Lung Cancer Cells via AKT Pathway. Onco Targets Ther 2020;13:3129-39. [Crossref] [PubMed]
- Song J, Jiang J, Wei N, et al. High CTSL2 expression predicts poor prognosis in patients with lung adenocarcinoma. Aging (Albany NY) 2021;13:22315-31. [Crossref] [PubMed]
- Ettinger DS. Ten years of progress in non-small cell lung cancer. J Natl Compr Canc Netw 2012;10:292-5. [Crossref] [PubMed]
- Yi H, Wang K, Jin JF, et al. Elevated Adenylyl Cyclase 9 Expression Is a Potential Prognostic Biomarker for Patients with Colon Cancer. Med Sci Monit 2018;24:19-25. [Crossref] [PubMed]
- Calebiro D, Grassi ES, Eszlinger M, et al. Recurrent EZH1 mutations are a second hit in autonomous thyroid adenomas. J Clin Invest 2016;126:3383-8. [Crossref] [PubMed]
- Kwanhian W, Lenze D, Alles J, et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med 2012;1:141-55. [Crossref] [PubMed]
- Lazar AM, Irannejad R, Baldwin TA, et al. G protein-regulated endocytic trafficking of adenylyl cyclase type 9. Elife 2020;9:e58039. [Crossref] [PubMed]
- Flanagan JM, Frohlich DM, Howard TA, et al. Genetic predictors for stroke in children with sickle cell anemia. Blood 2011;117:6681-4. [Crossref] [PubMed]
- Manjurano A, Clark TG, Nadjm B, et al. Candidate human genetic polymorphisms and severe malaria in a Tanzanian population. PLoS One 2012;7:e47463. [Crossref] [PubMed]
- Teixeira HM, Alcantara-Neves NM, Barreto M, et al. Adenylyl cyclase type 9 gene polymorphisms are associated with asthma and allergy in Brazilian children. Mol Immunol 2017;82:137-45. [Crossref] [PubMed]
- Rautureau Y, Deschambault V, Higgins MÈ, et al. ADCY9 (Adenylate Cyclase Type 9) Inactivation Protects From Atherosclerosis Only in the Absence of CETP (Cholesteryl Ester Transfer Protein). Circulation 2018;138:1677-92. [Crossref] [PubMed]
- Hopewell JC, Ibrahim M, Hill M, et al. Impact of ADCY9 Genotype on Response to Anacetrapib. Circulation 2019;140:891-8. [Crossref] [PubMed]
- Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [Crossref] [PubMed]
- The Gene Ontology (GO) project in. Nucleic Acids Res 2006;34:D322-6. [Crossref] [PubMed]
- Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482-517. [Crossref] [PubMed]
- Niu Y, Wang X, Peng Y. geecure: An R-package for marginal proportional hazards mixture cure models. Comput Methods Programs Biomed 2018;161:115-24. [Crossref] [PubMed]
- Tang Z, Shen Y, Zhang X, et al. The spike-and-slab lasso Cox model for survival prediction and associated genes detection. Bioinformatics 2017;33:2799-807. [Crossref] [PubMed]
- Cronin KA, Lake AJ, Scott S, et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer 2018;124:2785-800. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Feng Y, Sun W, Zhang J, et al. Neoadjuvant PD-1 inhibitor combines with chemotherapy versus neoadjuvant chemotherapy in resectable squamous cell carcinoma of the lung. Thorac Cancer 2022;13:442-52. [Crossref] [PubMed]
- Huang D, Cui P, Huang Z, et al. Anti-PD-1/L1 plus anti-angiogenesis therapy as second-line or later treatment in advanced lung adenocarcinoma. J Cancer Res Clin Oncol 2021;147:881-91. [Crossref] [PubMed]
- Isla D, de Castro J, García-Campelo R, et al. Treatment options beyond immunotherapy in patients with wild-type lung adenocarcinoma: a Delphi consensus. Clin Transl Oncol 2020;22:759-71. [Crossref] [PubMed]
- Kang DH, Jung SS, Yeo MK, et al. Suppression of Mig-6 overcomes the acquired EGFR-TKI resistance of lung adenocarcinoma. BMC Cancer 2020;20:571. [Crossref] [PubMed]
- Lou L, Wang J, Lv F, et al. Y-box binding protein 1 (YB-1) promotes gefitinib resistance in lung adenocarcinoma cells by activating AKT signaling and epithelial-mesenchymal transition through targeting major vault protein (MVP). Cell Oncol (Dordr) 2021;44:109-33. [Crossref] [PubMed]
- Saito R, Miki Y, Ishida N, et al. The Significance of MMP-1 in EGFR-TKI-Resistant Lung Adenocarcinoma: Potential for Therapeutic Targeting. Int J Mol Sci 2018;19:609. [Crossref] [PubMed]
- Seifert R, Lushington GH, Mou TC, et al. Inhibitors of membranous adenylyl cyclases. Trends Pharmacol Sci 2012;33:64-78. [Crossref] [PubMed]
- Hacker BM, Tomlinson JE, Wayman GA, et al. Cloning, chromosomal mapping, and regulatory properties of the human type 9 adenylyl cyclase (ADCY9). Genomics 1998;50:97-104. [Crossref] [PubMed]
- Iseki M, Park SY. Photoactivated Adenylyl Cyclases: Fundamental Properties and Applications. Adv Exp Med Biol 2021;1293:129-39. [Crossref] [PubMed]
- Halls ML, Cooper DMF. Adenylyl cyclase signalling complexes - Pharmacological challenges and opportunities. Pharmacol Ther 2017;172:171-80. [Crossref] [PubMed]
- Chang HM, Klausen C, Leung PC. Antimüllerian hormone inhibits follicle-stimulating hormone-induced adenylyl cyclase activation, aromatase expression, and estradiol production in human granulosa-lutein cells. Fertil Steril 2013;100:585-92.e1. [Crossref] [PubMed]
- Pálvölgyi A, Simpson J, Bodnár I, et al. Auto-inhibition of adenylyl cyclase 9 (AC9) by an isoform-specific motif in the carboxyl-terminal region. Cell Signal 2018;51:266-75. [Crossref] [PubMed]
- Niesor EJ, Benghozi R. Potential Signal Transduction Regulation by HDL of the β2-Adrenergic Receptor Pathway. Implications in Selected Pathological Situations. Arch Med Res 2015;46:361-71. [Crossref] [PubMed]
- Niesor EJ, Benghozi R, Amouyel P, et al. Adenylyl Cyclase 9 Polymorphisms Reveal Potential Link to HDL Function and Cardiovascular Events in Multiple Pathologies: Potential Implications in Sickle Cell Disease. Cardiovasc Drugs Ther 2015;29:563-72. [Crossref] [PubMed]
- Marsden AN, Dessauer CW. Nanometric targeting of type 9 adenylyl cyclase in heart. Biochem Soc Trans 2019;47:1749-56. [Crossref] [PubMed]
- Wu Y, Xia Y, Li P, et al. Role of the ADCY9 gene in cardiac abnormalities of the Rubinstein-Taybi syndrome. Orphanet J Rare Dis 2020;15:101. [Crossref] [PubMed]
- Li H, Liu Y, Liu J, et al. Assessment of ADCY9 polymorphisms and colorectal cancer risk in the Chinese Han population. J Gene Med 2021;23:e3298. [Crossref] [PubMed]
- Chao X, Jia Y, Feng X, et al. A Case-Control Study of ADCY9 Gene Polymorphisms and the Risk of Hepatocellular Carcinoma in the Chinese Han Population. Front Oncol 2020;10:1450. [Crossref] [PubMed]
- Zhang G, Xi M, Li Y, et al. The ADCY9 genetic variants are associated with glioma susceptibility and patient prognosis. Genomics 2021;113:706-16. [Crossref] [PubMed]
- Jia S, Li L, Xie L, et al. Transcriptome Based Estrogen Related Genes Biomarkers for Diagnosis and Prognosis in Non-small Cell Lung Cancer. Front Genet 2021;12:666396. [Crossref] [PubMed]
- Fang LT, Lee S, Choi H, et al. Comprehensive genomic analyses of a metastatic colon cancer to the lung by whole exome sequencing and gene expression analysis. Int J Oncol 2014;44:211-21. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Tan X, Banerjee P, Liu X, et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest 2018;128:3198. [Crossref] [PubMed]
- Ma Y, Zou H. Identification of the circRNA-miRNA-mRNA Prognostic Regulatory Network in Lung Adenocarcinoma. Genes (Basel) 2022;13:885. [Crossref] [PubMed]
- Wang Y, Hui J, Li R, et al. GBX2, as a tumor promoter in lung adenocarcinoma, enhances cells viability, invasion and migration by regulating the AKT/ERK signaling pathway. J Gene Med 2020;22:e3147. [Crossref] [PubMed]
- Zhou H, Zhang H, Shi M, et al. A robust signature associated with patient prognosis and tumor immune microenvironment based on immune-related genes in lung squamous cell carcinoma. Int Immunopharmacol 2020;88:106856. [Crossref] [PubMed]
- Song Q, Wang H, Bao J, et al. Systems biology approach to studying proliferation-dependent prognostic subnetworks in breast cancer. Sci Rep 2015;5:12981. [Crossref] [PubMed]
- Song Q, Zhu X, Jin L, et al. SMGR: a joint statistical method for integrative analysis of single-cell multi-omics data. NAR Genom Bioinform 2022;4:lqac056. [Crossref] [PubMed]