
PTMA binds to HMGB1 to regulate mitochondrial oxidative phosphorylation and thus affect the malignant progression of esophageal squamous cell carcinoma
Highlight box
Key findings
• Prothymosin alpha (PTMA) binds to high mobility group box 1 (HMGB1) to regulate mitochondrial oxidative phosphorylation, and thus, affects the malignant progression of esophageal squamous cell carcinoma (ESCC).
What is known and what is new?
• PTMA has been reported to serve as an oncogene in multiple malignancies. Importantly, PTMA is highly expressed in ESCC tissues and can can be used as a prognostic biomarker for ESCC.
• PTMA contributes to the progression of ESCC through binding to HMGB1 and regulating mitochondrial oxidative phosphorylation.
What are the implications, and what should change now?
• PTMA may be a critical and potential therapeutic target for ESCC, and this paper may provide a theoretical basis for the targeted therapy of ESCC.
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most common digestive tract malignancies worldwide. Esophageal cancer is usually divided into squamous cell carcinoma and adenocarcinoma according to histopathological characteristics, and ESCC accounts for about 90% of cases in China (1). Despite continuous improvements in the diagnosis and treatment of ESCC, the prognosis of patients remains very poor, and the 5-year survival rate is only 15–34% (2). A previous study has reported that the survival rate of patients with ESCC is mainly negatively correlated with tumor invasiveness and the presence of lymph node and distant metastases. Meanwhile, the occurrence and metastasis of ESCC are related to the overexpression of some oncogenes (3). Therefore, it is necessary to find new molecular biomarkers and therapeutic targets for ESCC.
Mitochondria are the main organelles of energy metabolism, and their basic function is oxidative phosphorylation (OXPHOS) through the electron transport chain (ETC) and adenosine triphosphate (ATP) synthetase, as well as providing cellular energy in the form of ATP (4). Mammalian cells can supply ATP through mitochondrial OXPHOS and glycolysis. Under normoxic conditions, mitochondrial OXPHOS is the main source of ATP in normal cell glucose metabolism, providing energy for metabolic activities (5). To maintain rapid and massive growth, tumor cells have increased metabolic and energy demands, which promotes the transformation of tumor cells by the Warburg effect or aerobic glycolysis (6). However, an increasing number of studies have found that mitochondrial OXPHOS also plays an important role in the progression of tumor malignancy. It has been reported that peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1α) can regulate mitochondrial function and OXPHOS to promote tumor metastasis (7). MYC proto-oncogene, bHLH transcription factor (MYC) and myeloid Cell Leukemia 1 (MCL1) can increase the level of mitochondrial OXPHOS, thereby enhancing the chemoresistance of breast cancer cells (8). Mitochondrial DNA content increases in ESCC tissues compared with normal tissues, indicating the activation of OXPHOS, which is negatively correlated with the prognosis of patients (9). Mitochondrial OXPHOS also plays an important role in some cancers, and OXPHOS may be a new target for anti-tumor therapy.
Prothymosin alpha (PTMA) is a nuclear protein containing 109–110 highly acidic amino acids, which is widely expressed in various cells and is involved in a variety of cellular biological behaviors, including the cell cycle, proliferation, apoptosis, gene transcription, and immune regulation (10). Recent studies have found that the expression of PTMA is closely related to the development of a variety of human malignant tumors, including bladder cancer, colon cancer, liver cancer, etc. (10-12). In hepatocellular carcinoma, PTMA is highly expressed, and reducing PTMA expression can induce Bax to enter mitochondria, promote the release of cytochrome C, and enhance the sensitivity of hepatocellular carcinoma cells to cisplatin (12). In colon cancer, PTMA expression can promote the proliferation of tumor cells, regulate the expression of the mitochondrial metabolic protein sterol regulatory element binding protein-1 (SREBP-1) through signal transducer and activator of transcription 3 (STAT3), and promote the resistance of colon cells to gemcitabine (10,13). Moreover, PTMA can also be used as a biomarker for ESCC, and is highly expressed in ESCC; the higher the expression level, the worse the prognosis (14,15). However, whether PTMA can affect tumor cell progression by regulating mitochondrial OXPHOS in ESCC remains unknown.
High mobility group box 1 (HMGB1) is a highly conserved nucleoprotein that has been uncovered to play a role in the pathogenesis of inflammatory diseases and malignant tumors (16,17). Further, HMGB1 overexpression has been reported to be associated with poorer overall survival rate and progression-free survival of ESCC patients and promote aggressive phenotypes and radioresistance of ESCC cells (18,19). However, the impacts of HMGB1 on oxidative phosphorylation in ESCC and the relationship between PTMA and HMGB1 have not been covered.
Therefore, in this paper, we discuss the role of PTMA in ESCC and its role and regulatory mechanism on mitochondrial OXPHOS in ESCC. Our paper provides a theoretical basis for the targeted therapy of ESCC. We present the following article in accordance with the MDAR and ARRIVE reporting checklists (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-143/rc).
Methods
Database
The ENCORI database (http://starbase.sysu.edu.cn/index.php) was used to analyze the expression level of PTMA in ESCC tissues and the overall survival rate of ESCC patients.
Patient tumor tissue collection
ESCC cancer tissues and para-carcinoma tissues were collected from 17 patients with ESCC who underwent initial surgery at the Fujian Medical University Union Hospital. Patients who received preoperative chemo- or radiotherapy were excluded from our study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fujian Medical University Union Hospital (approval No. 2022-198), and all participants have signed written informed consent.
Cell culture
ESCC cell lines (KYSE-30, KYSE150, KYSE410, and TE-1) obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were grown in 1640 media (Gibco, USA) medium supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin-streptomycin. Normal esophageal epithelial cells (HET-1A) obtained from the Cell Bank of the Chinese Academy of Sciences were grown in a bronchial epithelial growth medium (BEGM) kit medium (Lonza/Clonetics, Walkersville, MD, USA, CC-3170). All cells were in a 5% carbon dioxide (CO2) atmosphere at 37 ℃.
Nude mice xenograft model
Ten female BALB/c-nude mice (4–6 weeks old, Comparative Medicine Centre of Yangzhou University) (20,21) were supplied with free access to a standard rodent diet and tap water under temperature of 23±1 ℃ and a 12 h alternate light-dark cycle. The mice were divided into short hairpin-negative control (sh-NC) and sh-PTMA groups (n=5) using the random number table method. Briefly, the mice were anesthetized with an intraperitoneal injection of pentobarbital (50 mg/kg). No signs of peritonitis, pain, or discomfort were observed after anesthesia. Then the mice were subcutaneously injected with 5×106 KYSE30 cells treated with short hairpin RNA (shRNA) lentivirus into the right flank region of each mouse. The mice injected with KYSE30 cells transfected with sh-NC plasmid were referred to the negative control. Tumor volume was tested using the formula: length × width2/2 every 3 days following the formation of visible tumors to ensure compliance with humane and scientific endpoints. To minimize suffering, the total tumor burden did not exceed 10% of body weight, no tumors were allowed to grow larger than 20 mm in diameter and the mice were observed carefully for any signs of discomfort as judged by their behavior and food consumption. All mice were euthanized 21 days later by dislocating the cervical vertebrae, and tumor tissues were collected for related detection. All animal procedures were performed under a project license (approval No. IACUC-20220613-05) granted by the Institutional Animal Care and Use Committee of Jofunhwa Biotechnology (Nanjing) Co., Ltd., in compliance with institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Real-time quantitative reverse transcription-PCR (RT-qPCR)
qRT-PCR was performed on total RNA extracted with Trizol (Invitrogen, Carlsbad, CA, USA) and 200 nM of primers, and then isolated RNA was reverse-transcribed into complementary DNA (cDNA) using the PrimeScript RT reagent kit (Takara Biotechnology, Dalian, China). The thermal conditions of reverse transcription were as follows: 37 ℃ for 15 min and 85 ℃ for 5 sec. RT-PCR was performed with SYBR Green supermix (Takara Biotechnology, Dalian, China, RR420L) using a two-step PCR reaction procedure. The PCR conditions: 95 ℃ for 10 min for initial denaturation, 40 cycles of denaturation 15 sec at 95 ℃, annealing 30 sec at 60 ℃ and elongation 30 sec at 72 ℃ and final extension for 5 min at 72 ℃. The expression of PTMA and HMGB1 was normalized to the expression of β-actin. Relative quantification was calculated using the ΔΔCt method and normalized against β-actin (22). The following primers were used: PTMA, forward, 5'- TGAGGAAGAGGATGGAGATGAA-3'; reverse, 5'- CGGTCTTCTGCTTCTTGGTATC-3'; and HMGB1, forward, 5'- GGCCCGTTATGAAAGAGAAATG-3'; reverse, 5'- CTCAGAGCAGAAGAGGAAGAAG-3'; and β-actin, forward, 5'- AGCGAGCATCCCCCAAAGTT-3'; reverse, 5'- GGGCACGAAGGCTCATCATT-3'.
Western blot
Cell lysates were obtained using the radioimmunoprecipitation assay (RIPA) lysate on ice for 30 mins, and then the protein concentration was detected using a bicinchoninic acid (BCA) kit (Beyotime, Shanghai, China). Equal amounts (30 µg) of denatured protein lysates were resolved onto 15% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteins from the gel were transferred to a 0.2-µm polyvinylidene fluoride (PVDF) membrane. This was followed by blocking with 5% bovine serum albumin (BSA) for 1 hour. Subsequently, the blots were incubated with the following primary antibodies: anti-PTMA (1:1,000, abs147731, Absin, Shanghai, China), anti-HMGB1 (1:1,000, 66525-1-Ig, Proteintech, Rosemont, IL, USA), anti-B cell lymphoma-2 (Bcl-2; 1:1,000, 12789-1-AP, Proteintech, Rosemont, IL, USA), anti-Bax (1:1,000, 50599-2-Ig, Proteintech, Rosemont, IL, USA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:1,000, 0755R, Bioss, Beijing, China), which was followed by incubation with horseradish peroxidase (HRP)-Goat Anti-Rabbit immunoglobin G (IgG) (1:1,000, ab6721; Abcam) and HRP-Goat Anti-Mouse IgG secondary antibodies (1:1,000, ab6728; Abcam). Then, an Electrochemiluminescence (ECL) substrate (Pierce™ ECL Western Blotting Substrate, Thermo Fisher, MA, USA) was used to visualize the specific bands, and the dates were analyzed with Image J (National Institutes of Health, Bethesda, MD, United States).
Cell transfection
Gene silencing used short-hairpin RNA plasmid to express shRNA to silence PTMA (sh-PTMA#1, sh-PTMA#2, and sh-PTMA#3), viewing shRNA-NC as a negative control. Transfection of the shRNA plasmid designed by Shanghai GenePharma Co., Ltd. was conducted along with Lipofectamine 2000 (11668-027, Invitrogen, Carlsbad, CA, USA) in cell suspensions kept in six-well plates for 48 h. Target sequences for shRNA were as follows: shRNA-PTMA1: GTAGACGAAGAAGAGGAAGAA, shRNA-PTAM2: GAAGTTGTGGAAGAGGCAGAA, shRNA-PTAM3: GATGAGGATGACGATGTCGAT, and shRNA-NC: CCTAAGGTTAAGTCGCCCTCG. Overexpression of the HMGB1 (Ov-HMGB1) plasmid and overexpression (Ov)-NC plasmid were available from GenePharma (Shanghai, China) and the transfection was performed according to the protocol of the Lipofectamine 2000. Cells subjected to plasmid transfection were harvested for further use 48 h post transfection.
Cell Counting Kit-8 (CCK-8)
Cell proliferation was evaluated using a CCK-8 kit (Dojindo, Kumamoto, Japan). Briefly, cells were plated in 96-well plates at a density of 5×103 cells per well, and after the corresponding treatment, the cells were incubated in 10% CCK-8 reagent for another 2 h. The optical density (OD) value was measured at 450 nm with a microplate reader from Bio-Rad (Hercules, CA, USA).
5-ethynyl-2'-deoxyuridine (EdU) assay
In short words, 10 µM of EdU (Beyotime, Shanghai, China) was added to the cells inoculated in 12-well plates for 2 h after the corresponding treatment. Alexa Fluor 488 azide and 4',6-diamidino-2-phenylindole (DAPI) were employed to label the incorporated EdU and nuclear DNA for 30 min and 10 min following the 10 min of immobilization and 20 min of permeabilization of cells with 4% paraformaldehyde and 0.3% Triton X-100, respectively. Finally, ImageJ software was applied to analyze the percentage of EdU-positive cells photographed under a microscope (Leica, Germany, DMI8).
Cell apoptosis
A Cell Apoptosis Detection Kit with Annexin V-mCherry and SYTOX Green (Beyotime, Shanghai, China) was used for cell apoptosis detection as per the user manual. After the corresponding treatment, PBS rinse was performed in the collected cells. Prior to resuspension in 100 µL of binding buffer, the immobilization of cells with 70% ethanol was implemented for 1 h on ice. The PBS-rinsed cells were mixed with 400 µL of binding buffer at 4 ℃ for 20 min after 15 min of incubation of Annexin V-mCherry and SYTOX Green under light-proof conditions. Lastly, the apoptosis rate was subjected to analysis with a flow cytometer (FACSCanto II; BD Biosciences, San Jose, CA).
Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay
Reactive oxygen species (ROS) production was detected using ROS-specific fluorescent probe 2', 7'-Dichlorofluorescin diacetate (DCFH-DA, Beyotime, Shanghai, China) as per the user manual. After the corresponding treatment, the cells seeded in six-well plates (5×106 cells/well) were incubated with 10 µM of DCFH-DA for 30 min at 37 ℃ in dark. Then, cells were washed and examined using a microplate reader from Bio-Rad and Olympus fluorescence microscopy (Tokyo, Japan).
Mitochondrial ROS assessment
Mitochondrial ROS levels were detected using the fluorescent probes MitoSOX™ Red (Molecular Probes, Life Technologies, Carlsbad, CA, USA) as per the user manual, and fluorescence intensity was measured using a flow cytometer (FACSCanto II; BD Biosciences, San Jose, CA).
Mitochondrial Membrane Potential (MMP) Assay
MMP was assayed using 5,5',6,6'-tetrachloro-1,1',3,3'-tetraethyl-benzimidazolyl carbocyanine iodide (JC-1) as per the user manual (Beyotime Biotech, Nanjing, China). Briefly, After the corresponding treatment, the cells seeded in six-well plates (5×106 cells/well) were stained with JC-1 and were analyzed using a flow cytometer (FACSCanto II; BD Biosciences, San Jose, CA) at a wavelength of 488 nm. Also, the approximate emission wavelengths of the monometric and J-aggregate forms are 529 and 590 nm.
The activities of mitochondrial respiratory chain complexes I, III, and IV
The cells were plated in 96-well plates at a density of 5×103 cells per well, and after the corresponding treatment, the cells were collected and mitochondrial respiratory chain complexes I, III, IV detection kits (LabTech, Beijing, China) were used to detect the activity of the complex.
ATP assay
After the corresponding treatment, the cells were seeded in six-well plates (5×106 cells/well) and the ATP Bioluminescence Assay Kit (Sigma-Aldrich, St. Louis, MO, USA) was used to detect the ATP level as per the user manual.
Co-immunoprecipitation (co-IP)
The treated cells seeded in six-well plates (5×106 cells/well) were probed with FLAG-tagged PTMA or hemagglutinin (HA)-tagged HMGB1 at 4 ℃ for 3 h following the addition of lysis buffer and centrifugation at 300 g for 10 min. Then, immunoprecipitated proteins were analyzed following the supplementation of 40 µL of sample buffer.
Immunofluorescence (IF)
Following immobilization and permeabilization of the cells with 4% paraformaldehyde and 0.1% Triton X-100 for 15 min, respectively, the overnight cultivation of the cells impeded by 10% goat serum with PTMA (1:300) and HMGB1 (1:300) antibodies was carried out at 4 ℃, followed by the exposure to Alexa Fluor®594 and Alexa Fluor® 488-conjugated secondary antibodies (1:400; Abcam, Cambridge, UK) for 90 min. Finally, Hoechst (Beyotime) staining lasted for 5 min. The distribution of PTMA and HMGB1 were tracked using the Olympus fluorescence microscope.
Immunohistochemistry (IHC)
After dewaxing, rehydration, retrieval of antigens, the paraffin-embedded ESCC tissue sections impeded by 0.05% BSA were subjected to the cultivation with anti-Ki67 and anti-PTMA overnight at 4 ℃, prior to the exposure to the secondary antibodies for 1 h and 10 min of DAPI staining at room temperature. Fluorescein isothiocyanate (FITC)-labeled wheat-germ-agglutinin (WGA, Sigma-Aldrich, St. Louis, MO, USA) was used to visualize and quantify the cross-sectional area of the tumor tissues with ImageJ software.
Statistical analysis
All experiments were independently repeated in triplicate and all experimental data were biologically repeated in triplicate. GraphPad Prism 7 (GraphPad Software, San Diego, California USA) was employed to analyze the data. Comparisons were performed using the Student’s t-test or one-way analysis of variance (ANOVA) analysis. P<0.05 was regarded as statistically significant.
Results
The expression of PTMA in ESCC is significantly elevated
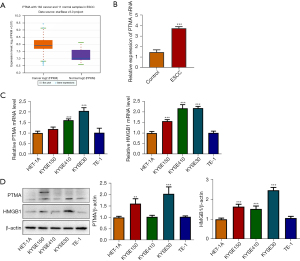
The ENCORI database showed that PTMA expression was significantly increased in ESCC tissues (Figure 1A). PTMA expression in ESCC tumor tissues of clinical patients was detected by RT-qPCR, and the results showed that compared with the control group, the expression of PTMA in ESCC group was significantly increased (Figure 1B). Subsequently, the expression of PTMA and HMGB1 in ESCC cells was detected by RT-qPCR and Western blot. The results showed that compared with HET-1A cells, the expressions of PTMA and HMGB1 in ESCC cell lines (KYSE30, KYSE150, KYSE410, and TE-1) were also significantly increased (Figure 1C,1D). The expression of PTMA in KYSE-30 cells was most significantly increased, so KYSE-30 was selected for the follow-up experiment.
Interference with PTMA expression inhibits ESCC cell proliferation and promotes cell apoptosis
The expression of PTMA in KYSE-30 cells was inhibited, and the cells were divided into control, sh-NC, sh-PTMA#1, sh-PTMA#2, and sh-PTMA#3 groups. The RT-qPCR and Western blot results showed that compared with the sh-NC group, the expression of PTMA in the sh-PTMA#1, sh-PTMA#2, and sh-PTMA#3 groups was significantly decreased, and the decrease was most significant in the sh-PTMA#3 group. Therefore, sh-PTMA#3 was selected for the follow-up experiments (Figure 2A,2B).
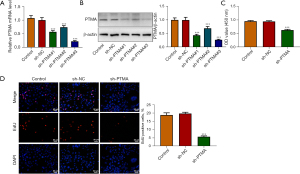
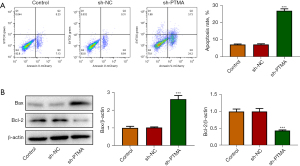
The CCK8 results delineated that the cell activity following PTMA depletion was significantly diminished (Figure 2C). The EdU staining results disclosed that relative to the sh-NC group, the cell proliferation ability in the sh-PTMA group was significantly weakened (Figure 2D). The flow cytometry and Western blot results showed that by contrast with sh-NC, the apoptotic rate was significantly potentiated after PTMA silencing, along with increased expression of Bax and decreased expression of Bcl-2 (Figure 3A,3B).
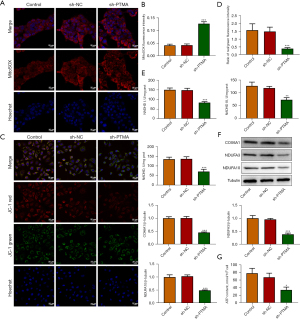
Interference with PTMA causes ROS aggregation in ESCC cells by inhibiting mitochondrial oxidative phosphorylation
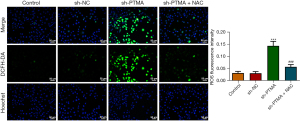
DCFH-DA was used to detect intracellular ROS levels, and the results showed that ROS level was significantly increased in the sh-PTMA group compared with the sh-NC group. Further administration of the ROS inhibitor N-acetylcysteine (NAC) reversed ROS level in cells (Figure 4), suggesting that interference with PTMA caused ROS aggregation in ESCC cells. Subsequently, the MitoSOX fluorescent probe was used to detect mitochondrial ROS levels, and the results showed that the level of mitochondrial ROS was significantly increased after PTMA inhibition (Figure 5A,5B). Relative to sh-NC, MMP expression after PTMA absence was markedly decreased by JC-1 staining (Figure 5C,5D). The inhibition of PTMA expression in ESCC cells significantly decreased the expression of mitochondrial complexes I, II, and III (Figure 5E). Western blot detection of the mitochondrial energy metabolism-related proteins illuminated that NDUFA3, NDUFA10, and COX6A1 expressions were notably decreased when PTMA was lowly expressed (Figure 5F). ATP level in cells was detected by ATP kit, and the results showed that ATP level was remarkably depleted by PTMA deficiency (Figure 5G).
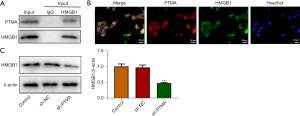
PTMA is combined with HMGB1
The co-IP results showed that the expression of PTMA in cells was significantly decreased after co-precipitation of HMGB1 (Figure 6A). The co-localization of PTMA and HMGB1 was detected by IF, and the results showed that PTMA and HMGB1 were both expressed in both the nucleus and cytoplasm, and were co-expressed in the cells (Figure 6B). Western blot was used to detect the expression level of HMGB1 after PTMA interference, and the results showed that the expression of HMGB1 in cells was significantly decreased following inhibition of PTMA expression (Figure 6C).
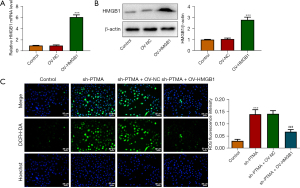
The binding of PTMA to HMGB1 affects intracellular ROS levels and the expression of mitochondrial respiratory chain complexes
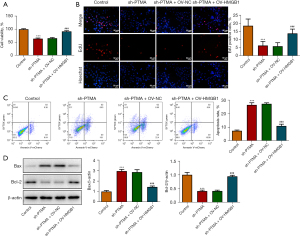
We then overexpressed HMGB1 in the cells and detected the transfection efficiency by Western blot and RT-qPCR (Figure 7A,7B). Next, we divided the cells into control, sh-PTMA, sh-PTMA+OV-NC, and sh-PTMA+OV-HMGB1 groups. The DCFH-DA test results showed that ROS level in the sh-PTMA+OV-HMGB1 group was prominently lessened relative to that in the sh-PTMA+OV-NC group (Figure 7C). The MitoSOX fluorescence probe results showed that mitochondrial ROS level in the sh-PTMA+OV-HMGB1 group was significantly decreased compared with the sh-PTMA+OV-NC group (Figure 8A,8B). JC-1 staining corroborated that relative to the sh-PTMA+OV-NC group, MMP was significantly increased by HMGB1 elevation (Figure 8C,8D), accompanied by increased expression of mitochondrial complexes I (Figure 8E). The Western blot results showed that the overexpression of HMGB1 could significantly reverse the inhibitory effect of PTMA interference on mitochondria-related proteins (Figure 8F). Also, the ATP detection results showed that overexpression of HMGB1 could significantly reverse the inhibitory effect of PTMA interference on ATP (Figure 8G).
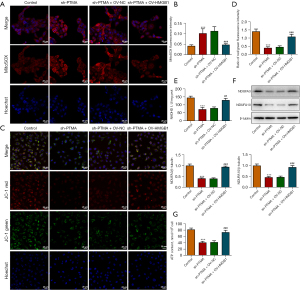
PTMA binding to HMGB1 affects the proliferation and apoptosis of ESCC cells
The CCK8 and EdU staining results substantiated that relative to the sh-PTMA+OV-NC group, the cell activity and proliferation ability were distinctly increased by HMGB1 overexpression (Figure 9A,9B). The flow cytometry results showed that HMGB1 overexpression could significantly reverse the inhibitory effect of PTMA interference on apoptosis (Figure 9C). Furthermore, the Western blot results showed that compared with the sh-PTMA+OV-NC group, Bax expression was inhibited and Bcl-2 expression was increased in the sh-PTMA+OV-HMGB1 group (Figure 9D).
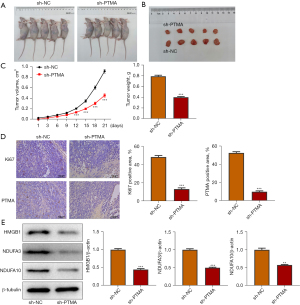
Interference with PTMA expression inhibits the proliferation of tumor tissue in ESCC mice
BALB/c nude mice were divided into sh-NC and sh-PTMA, and Figure 10A displays a picture of the mice. Figure 10B is a photograph of mouse tumors. The tumor volume and tumor weight results presented that relative to the sh-NC group, the tumor volume and tumor weight of mice in the sh-PTMA group were evidently reduced (Figure 10C). The expressions of PTMA and the proliferation-related protein Ki67 in tissues were detected by IHC, and the results demonstrated that the expressions of Ki67 and PTMA were significantly decreased following the inhibition of PTMA expression (Figure 10D). Western blot results showed that compared with the sh-NC group, the expressions of HMGB1, NDUFA3, and NDUFA10 in tumor tissues of the sh-PTMA group were significantly decreased (Figure 10E).
Discussion
According to a World Health Organization global cancer (GLOBOCAN) 2018 study, 572,000 people were newly diagnosed with ESCC worldwide in 2018 and 509,000 people died from the disease. ESCC is the most common esophageal cancer in East and Central Asia, South Africa, and Africa, accounting for about 90% of esophageal cancer cases globally (23). Therefore, there is a pressing need to study the pathogenesis of ESCC to improve the therapeutic effect of patients with ESCC and explore new prognostic targets.
Oxidative stress is a reaction to the accumulation of ROS and the imbalance of intracellular redox ability, which affects a variety of biological behaviors of cells, including oxidative stress injury as well as cell proliferation and apoptosis after injury (24). In the tumor environment, ROS production is closely related to tumorigenesis, metabolism, invasiveness, and drug resistance (25). Tumor cells exhibit higher levels of ATP and ROS production than normal cells to maintain their higher proliferation and invasion abilities (26). However, excessive ROS can also be cytotoxic to tumor cells (27). For example, the antitumor drugs, 5-fluorouracil and oxaliplatin, inhibit tumor cell division by inducing increased ROS production and interfering with DNA replication (28). Paclitaxel can increase the activity of nicotinamide adenine dinucleotide phosphate (NADPH) and produce more ROS, thus inhibiting the proliferation of tumor cells (29). However, these chemotherapy drugs not only target tumor cells but also damage normal cells, so it is necessary to find tumor cell-specific targets. Tetrahydrobenzimidazole TMQ0153 can induce ROS aggregation and lead to the death of chronic leukemia cells, releasing HMGB1 extracellularly (30). Reducing the expression of interferon-inducible protein 6 (IFI6) can cause mitochondrial dysfunction, promote ROS accumulation and inhibit the growth of esophageal squamous cell carcinoma (31). Therefore, specifically increasing ROS can selectively kill tumor cells (32).
The function of PTMA is closely related to cellular immune regulation, cell proliferation, and apoptosis. Since PTMA is a typical tumor-related protein, an in-depth study on its regulation of cell proliferation and apoptosis is helpful to discover the mechanism of cell carcinogenesis. Elevated PTMA levels have been detected in a variety of cancers (12,33). High levels of PTMA protein product in cancer cells not only maintain a highly proliferative state but also protect cells from entering the apoptotic pathway (34). In our experiment, PTMA expression was significantly increased in both the ESCC cells and patient tissues. In the cell experiments, we further discussed the regulatory role of PTMA in ESCC. We inhibited the expression of PTMA in ESCC cells and found that the proliferation and activity of ESCC cells were significantly decreased, while the apoptosis of ESCC cells was significantly increased. The results indicated that the inhibition of PTMA could inhibit the growth of ESCC tumor cells.
In addition, studies have shown that the expression of PTMA is affected during the cross-action between oxidative stress and DNA replication (35,36). However, whether PTMA regulates mitochondrial oxidative stress in tumor cells has not yet been reported. Therefore, we detected a correlation between mitochondrial oxidative phosphorylation and PTMA expression in ESCC cells. Our results showed that PTMA inhibition significantly increased the level of ROS in cells, mitochondrial ROS, and decreased MMP levels. The electron transport chain (ETC) is an electron transport orbit composed of respiratory transmitters on the inner mitochondrial membrane, containing complexes I, II, III, and IV, and exerts a major function in oxidative phosphorylation. It also plays a key role in the proliferation, survival, and metastasis of cancer cells (37). Complex I mainly consists of seven NADH dehydrogenase (ND) subunits (ND1-6 and ND4L) and 37 nuclear gene subunits (NDUFV1-3, NDUFS1-8, NDUFA1-13, NDUFB1-11, NDUFAB1, NDUFC1-2) encoded by mitochondrial DNA. It is one of the most important protein complexes for electrons to enter the respiratory chain, which can transfer electrons from nicotinamide adenine dinucleotide (NADH) to coenzyme Q (CoQ), and at the same time conjugate four protons from the mitochondrial matrix to pump out to the membrane gap, forming a transmembrane proton gradient, driving the synthesis of ATP, and promoting the tumor (38). As a by-product of oxidative metabolism, ROS is also released through the ETC. Studies have shown that the absence of OXPHOS complexes I, II, and III can lead to the increase of mitochondrial ROS and induce the apoptosis of tumor cells (39,40). Targeting the mitochondrial respiratory chain complex I and Bcl-2 inhibits the progression of oxidative phosphorylation-dependent acute leukemia (41). The down-regulation of oxidative phosphorylated complex I and IV and related proteins NDUFA1 and COX6B1 can inhibit the proliferation of ESCC both in vitro and in vivo (42). In our experiment, it was found that the inhibition of PTMA significantly increased the levels of ROS and mitochondrial ROS in cells, and was accompanied by a significant decrease in the expression of mitochondrial complexes I, II, and III, and a significant decrease in the expression of the mitochondria-related proteins, NDUFA3, NDUFA10, and COX6A1. Therefore, our experiment indicated that interference with PTMA could significantly inhibit mitochondrial oxidative phosphorylation in ESCC, thus inhibiting the growth of ESCC cells.
Database analysis showed that there was a protein interaction between HMGB1 and PTMA. HMGB1 is widely found in the cytoplasmic membrane, intracellular nucleus, cytoplasm, mitochondria, and other organelles, which can maintain nuclear homeostasis and is also a key regulator of mitochondrial function and morphology (43). Defects in the mitochondrial respiratory chain and ATP synthesis can be observed through HMGB1 targeted deletion (44). In tumor cells, the increase of HMGB1 can activate the activity of the respiratory chain complex I, regulate mitochondrial function, and promote the growth of pancreatic tumors (45). In this study, PTMA was found to bind to HMGB1, regulate the expression of the mitochondrial respiratory chain complex and related proteins, inhibit cell oxidative phosphorylation, and induce ROS aggregation, thereby inhibiting ESCC proliferation and migration and inducing ESCC cell apoptosis.
Conclusions
PTMA binds to HMGB1 to regulate mitochondrial oxidative phosphorylation and thus affects the malignant progression of ESCC.
Acknowledgments
Funding: The study was supported by Quanzhou City Science & Technology Program of China (No. 2022NS055).
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-143/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-143/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-143/coif). All authors report that they received funding from Quanzhou City Science & Technology Program of China (No. 2022NS055). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fujian Medical University Union Hospital (approval No. 2022-198), and all participants have signed written informed consent. All animal procedures were performed under a project license (approval No. IACUC-20220613-05) granted by the Institutional Animal Care and Use Committee of Jofunhwa Biotechnology (Nanjing) Co., Ltd., in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Malhotra GK, Yanala U, Ravipati A, et al. Global trends in esophageal cancer. J Surg Oncol 2017;115:564-79. [Crossref] [PubMed]
- Moral Moral GI, Viana Miguel M, Vidal Doce Ó, et al. Postoperative complications and survival rate of esophageal cancer: Two-period analysis. Cir Esp 2018;96:473-81. (Engl Ed). [Crossref] [PubMed]
- Xiao S, Huang G, Zeng W, et al. Roles of oncogenes in esophageal squamous cell carcinoma and their therapeutic potentials. Clin Transl Oncol 2023;25:578-91. [Crossref] [PubMed]
- Tian C, Liu Y, Li Z, et al. Mitochondria Related Cell Death Modalities and Disease. Front Cell Dev Biol 2022;10:832356. [Crossref] [PubMed]
- Warburg O, Wind F, Negelein E. THE METABOLISM OF TUMORS IN THE BODY. J Gen Physiol 1927;8:519-30. [Crossref] [PubMed]
- Di Gregorio J, Petricca S, Iorio R, et al. Mitochondrial and metabolic alterations in cancer cells. Eur J Cell Biol 2022;101:151225. [Crossref] [PubMed]
- LeBleu VS, O'Connell JT, Gonzalez Herrera KN, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 2014;16:992-1003, 1-15.
- Lee KM, Giltnane JM, Balko JM, et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab 2017;26:633-647.e7. [Crossref] [PubMed]
- Li H, Tian Z, Zhang Y, et al. Increased copy number of mitochondrial DNA predicts poor prognosis of esophageal squamous cell carcinoma. Oncol Lett 2018;15:1014-20. [PubMed]
- Jin L, Zhu LY, Pan YL, et al. Prothymosin α promotes colorectal carcinoma chemoresistance through inducing lipid droplet accumulation. Mitochondrion 2021;59:123-34. [Crossref] [PubMed]
- Hu CY, Su BH, Lee YC, et al. Interruption of the long non-coding RNA HOTAIR signaling axis ameliorates chemotherapy-induced cachexia in bladder cancer. J Biomed Sci 2022;29:104. [Crossref] [PubMed]
- Lin YT, Liu YC, Chao CC. Inhibition of JNK and prothymosin-alpha sensitizes hepatocellular carcinoma cells to cisplatin. Biochem Pharmacol 2016;122:80-9. [Crossref] [PubMed]
- Yang L, Sun H, Liu X, et al. Circular RNA hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-512-5p to upregulate the expression of PTMA. J Cell Physiol 2020; Epub ahead of print. [Crossref] [PubMed]
- Shao M, Li W, Wang S, et al. Identification of key genes and pathways associated with esophageal squamous cell carcinoma development based on weighted gene correlation network analysis. J Cancer 2020;11:1393-402. [Crossref] [PubMed]
- Zhu Y, Qi X, Yu C, et al. Identification of prothymosin alpha (PTMA) as a biomarker for esophageal squamous cell carcinoma (ESCC) by label-free quantitative proteomics and Quantitative Dot Blot (QDB). Clin Proteomics 2019;16:12. [Crossref] [PubMed]
- Zhang Y, Dong Q, Liu C, et al. Resveratrol affects the migration and apoptosis of monocytes by blocking HMGB1/NF-κB pathway. Transl Cancer Res 2021;10:3647-58. [Crossref] [PubMed]
- Wang S, Zhang Y. HMGB1 in inflammation and cancer. J Hematol Oncol 2020;13:116. [Crossref] [PubMed]
- Wu T, Zhang W, Yang G, et al. HMGB1 overexpression as a prognostic factor for survival in cancer: a meta-analysis and systematic review. Oncotarget 2016;7:50417-27. [Crossref] [PubMed]
- Dong J, Zhang X, Du X, et al. HMGB1 overexpression promotes a malignant phenotype and radioresistance in ESCC. J Cancer 2022;13:2717-26. [Crossref] [PubMed]
- Jia Y, Tian C, Wang H, et al. Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin. Mol Cancer 2021;20:162. [Crossref] [PubMed]
- Wu Q, Zhang W, Wang Y, et al. MAGE-C3 promotes cancer metastasis by inducing epithelial-mesenchymal transition and immunosuppression in esophageal squamous cell carcinoma. Cancer Commun (Lond) 2021;41:1354-72. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhang B, Pan C, Feng C, et al. Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep 2022;27:45-52. [Crossref] [PubMed]
- Cheung EC, Vousden KH. The role of ROS in tumour development and progression. Nat Rev Cancer 2022;22:280-97. [Crossref] [PubMed]
- Ishikawa K, Takenaga K, Akimoto M, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008;320:661-4. [Crossref] [PubMed]
- Hyun DH. Insights into the New Cancer Therapy through Redox Homeostasis and Metabolic Shifts. Cancers (Basel) 2020;12:1822. [Crossref] [PubMed]
- Afzal S, Jensen SA, Sørensen JB, et al. Oxidative damage to guanine nucleosides following combination chemotherapy with 5-fluorouracil and oxaliplatin. Cancer Chemother Pharmacol 2012;69:301-7. [Crossref] [PubMed]
- Alexandre J, Hu Y, Lu W, et al. Novel action of paclitaxel against cancer cells: bystander effect mediated by reactive oxygen species. Cancer Res 2007;67:3512-7. [Crossref] [PubMed]
- Song S, Lee JY, Ermolenko L, et al. Tetrahydrobenzimidazole TMQ0153 triggers apoptosis, autophagy and necroptosis crosstalk in chronic myeloid leukemia. Cell Death Dis 2020;11:109. [Crossref] [PubMed]
- Liu Z, Gu S, Lu T, et al. IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. J Exp Clin Cancer Res 2020;39:144. [Crossref] [PubMed]
- Idelchik MDPS, Begley U, Begley TJ, et al. Mitochondrial ROS control of cancer. Semin Cancer Biol 2017;47:57-66. [Crossref] [PubMed]
- Kumar A, Kumar V, Arora M, et al. Overexpression of prothymosin-α in glioma is associated with tumor aggressiveness and poor prognosis. Biosci Rep 2022;42:BSR20212685. [Crossref] [PubMed]
- Jiang G, Yu H, Li Z, et al. lncRNA cytoskeleton regulator reduces non small cell lung cancer radiosensitivity by downregulating miRNA 206 and activating prothymosin α. Int J Oncol 2021;59:88. [Crossref] [PubMed]
- Radulovic M, Baqader NO, Stoeber K, et al. Spatial Cross-Talk between Oxidative Stress and DNA Replication in Human Fibroblasts. J Proteome Res 2016;15:1907-38. [Crossref] [PubMed]
- Pinto G, Radulovic M, Godovac-Zimmermann J. Spatial perspectives in the redox code-Mass spectrometric proteomics studies of moonlighting proteins. Mass Spectrom Rev 2018;37:81-100. [Crossref] [PubMed]
- Greene J, Segaran A, Lord S. Targeting OXPHOS and the electron transport chain in cancer; Molecular and therapeutic implications. Semin Cancer Biol 2022;86:851-9. [Crossref] [PubMed]
- Cogliati S, Calvo E, Loureiro M, et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature 2016;539:579-82. [Crossref] [PubMed]
- Jang S, Javadov S. Elucidating the contribution of ETC complexes I and II to the respirasome formation in cardiac mitochondria. Sci Rep 2018;8:17732. [Crossref] [PubMed]
- Maranzana E, Barbero G, Falasca AI, et al. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal 2013;19:1469-80. [Crossref] [PubMed]
- Liu F, Kalpage HA, Wang D, et al. Cotargeting of Mitochondrial Complex I and Bcl-2 Shows Antileukemic Activity against Acute Myeloid Leukemia Cells Reliant on Oxidative Phosphorylation. Cancers (Basel) 2020;12:2400. [Crossref] [PubMed]
- Li B, Yu Y, Jiang Y, et al. Cloperastine inhibits esophageal squamous cell carcinoma proliferation in vivo and in vitro by suppressing mitochondrial oxidative phosphorylation. Cell Death Discov 2021;7:166. [Crossref] [PubMed]
- Wang D, Liu K, Fukuyasu Y, et al. HMGB1 Translocation in Neurons after Ischemic Insult: Subcellular Localization in Mitochondria and Peroxisomes. Cells 2020;9:643. [Crossref] [PubMed]
- Tang D, Kang R, Livesey KM, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab 2011;13:701-11. [Crossref] [PubMed]
- Kang R, Tang D, Schapiro NE, et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 2014;33:567-77. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


