
Comprehensive bioinformatics analysis on exportins in lung adenocarcinoma and lung squamous cell carcinoma
Highlight box
Key findings
• The transcriptional and protein expression levels of CSE1L and XPO1/5/6/7 were increased in patients with LUAD and LUSC, and the increased transcriptional levels of CSE1L and XPO5/6/7 were related to worse prognosis. The increased transcriptional level of XPO1 suggested a better prognosis. These results indicated that CSE1L and XPO1/5/6/7 potential prognostic biomarkers for the survival of patients with LUAD and LUSC.
What is known and what is new?
• Exportins are associated with cell activity and progression of various cancers.
• To analyze the differential expression, prognostic value, genetic variation, biological function, and immune cell infiltration of exportins in patients with LUAD and LUSC, the ONCOMINE; UALCAN; HPA, Kaplan-Meier plotter; cBioPortal; STRING; DAVID; TIMER; and LinkedOmics databases were used in this study.
What is the implication, and what should change now?
• Our study provides novel insights into the selection of prognostic biomarkers of exportins in LUAD and LUSC.
Introduction
As one of the most common malignant tumors in the world and the most common cause of global cancer-related mortality, lung cancer results in more than a million deaths each year (1). There are two main types of lung cancer, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), with the most common subtypes of NSCLC being lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), accounting for about 85% of new lung cancer cases (2). Despite the progress of molecular targeted and immune-based checkpoint therapies, the 5-year overall survival (OS) of patients with NSCLC remains less than 15% (3), and cancer therapy still faces great challenges, including delayed diagnosis, recurrence, metastasis, and cancer-related mutations. Therefore, it is crucial to determine more reliable tumor molecular markers for early the screening, diagnosis, individualized treatment, and prognosis of NSCLC.
Exportins are a group of nucleocytoplasmic transport receptor proteins that are widely distributed in eukaryotes. At present, 7 exportins have been identified in the mammalian cell, namely XPO1 (also called CRM1), CSE1L (also called XPO2 or CAS), XPOT (also called XPO3), XPO4, XPO5, XPO6, and XPO7 (4,5). These exportins belong to the Karyopherin-β (Kap-β; also known as importin and exportin) family and are responsible for transporting most proteins and RNA in the nucleus across the nuclear membrane to the cytoplasm (5,6). Kaps have low sequence identity (10–20%), share similar molecular weights (90–150 kDa) and isoelectric points (PI =4.0–5.0), and all have helical HEAT repeats. Through these special structures, biological macromolecules can be exported to the nucleus (7). Kaps are also closely linked to many cellular processes, including gene differential expression, cell signal transduction, tumor immune response, and tumorigenesis because they interact with a large number of proteins with different functions and play a role in controlling a variety of protein localizations (8). Some studies have found that exportins are associated with cell activity and progression of various cancers, such as liver cancer (9), breast cancer (10), gastric cancer (11), lung cancer (12), and prostate cancer (13). A study showed that CRM1 is frequently over-expressed in NSCLC, especially in LUAD and LUSC. Furthermore, the study also found that after tobacco carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) exposure or transfection with CRM1 vector, the overexpression of CRM1 in the lung epithelial cell line BEAS-2B led to cellular transformation, which suggests that the up-regulation of CRM1 may be an important pathway for malignant transformation of lung epithelial cells (14). CSE1L interacts with p65 and activates nuclear factor-κB (NF-κB), and Mitogen-activated protein kinase (MAPK) signaling pathway promotes NSCLC proliferation and inhibits apoptosis (15). However, the expression level, genetic variation, immune infiltration and biological function of different exportins in LUAD and LUSC was well as their relationship with prognosis of patients with LUAD and LUSC have not been fully clarified.
With the rapid development of RNA-sequencing technology and microarrays, RNA and DNA exploration have become a significant constituent of biomedical and biological research (16). For example, Guo et al. (17) analyzed the DEGs and hub genes that affect the development of LUAD through bioinformatics technology, providing potential diagnosis and treatment strategies for the treatment of LUAD. In our study, we used various large public databases to expand the relevant knowledge of LUAD and LUSC, and conducted a comprehensive bioinformatics analysis on the relationship between 7 different exportins and the prognosis of LUAD and LUSC. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-228/rc).
Methods
ONCOMINE analysis
ONCOMINE (www.ONCOMINE.org) is currently the world’s largest publicly accessible oncogene chip database and comprehensive data mining platform, including 715 data sets and 86,733 samples (18). The ONCOMINE database was used to determine the transcriptional levels of exportins in different types of cancer. In our study, a P value <1E-4, a fold change of 2, and a gene rank in the top 10% were set as the significance thresholds. A t-test was used to analyze the differences in the expression of exportins in LUAD and LUSC, and statically significant differences were considered present with a P value <0.05. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
UALCAN analysis
UALCAN (http://ualcan.path.uab.edu/) mainly uses The Cancer Genome Atlas (TCGA) transcriptome and clinical patient data to help identify candidate biomarkers of specific cancer subclasses with diagnostic, prognostic, or therapeutic significance, and it can also be used as a computer verification platform for target genes (19). In our study, the “TCGA analysis” module and the “LUAD” and “LUSC” data sets were used to analyze the relationship between exportins transcription levels in LUAD and LUSC and normal tissues in the “expression” link. Statically significant differences were considered present with a P value <0.05.
Human Protein Atlas analysis
The Human Protein Atlas (HPA; https://www.proteinatlas.org/) database is an online free database that provides a large amount of proteomic and transcriptomic data of specific human tissues and cells (20). In our study, the protein expression levels of exportins in LUAD and LUSC and normal tissues were studied.
Kaplan-Meier plotter analysis
As an important prognostic biomarker evaluation tool, the Kaplan-Meier plotter (https://kmplot.com/analysis/index.php?p=background) can evaluate the correlation between the expression of the 54,000 genes on the survival rates in 21 different cancers (21). We used this database to study the prognostic value of exportins mRNA expression in lung cancer, including overall survival (OS) and progression-free survival (PFS). We also evaluated the hazard ratio (HR) with 95% confidence intervals and log rank p value. Statically significant differences were considered present with a P value <0.05.
cBioPortal analysis
cBioPortal (https://www.cbioportal.org/) is a comprehensive network resource that can visualize and analyze multidimensional cancer genomics data (22). In our study, by using the cBioPortal online tool of LUAD (TCGA, Firehose legacy), the genome profiles of 7 exportin members were analyzed, which contained mutations, putative copy number alterations (CNAs) from genomic identification of significant targets in cancer (GISTIC), and mRNA expression Z scores (microarray). Coexpression genes of exportin members were determined using the “co-expression” module of cBioPortal. Pearson correlation coefficient was used to calculate the correlation between exportin members and coexpression genes, and the top 10 coexpression genes of each exportin with the largest Pearson correlation coefficient were identified.
Protein-protein interaction (PPI) network construction and module analysis
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; version 11.5; https://cn.string-db.org/) is an online database for analyzing the functional interactions between proteins encoded by different genes (23). Cytoscape (version 3.8.2) is an open bioinformatics software platform for visualizing gene interaction networks (24). The STRING database was used to construct the PPI network of 69 genes significantly related to exportins mutations, and then the module analysis was conducted in Cytoscape software.
Database for Annotation, Visualization, and Integrated Discovery analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID; version 6.8; https://david.ncifcrf.gov/tools.jsp) is a comprehensive bioinformatics website that can visually annotate biological functions (25). In our study, 69 coexpression genes significantly related to exportins mutations were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Statically significant differences were considered present at a P value <0.05.
Tumor Immune Estimation Resource analysis
Tumor Immune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer) is a reliable and convenient online analysis tool to analyze immune infiltration systematically in various malignancies. We used the “gene” module of this database to study the correlation between the expression of exportins in LUAD and LUSC and immune cell infiltration. A statically significant difference was considered present with a P value <0.05.
LinkedOmics analysis
LinkedOmics (http://www.linkedomics.org/login.php) is a comprehensive multiomics database for the analysis of 32 TCGA cancer types (26). We studied the microRNA (miRNA) target enrichment and transcription factor target enrichment of exportins through the “LinkInterpriser” module in TCGA_LUAD data set. Statically significant differences were considered present at a P value <0.05.
Results
Transcription levels and protein expression of exportins in patients with LUAD and LUSC
The transcriptional and protein levels of different exportins members between LUAD, LUSC, and normal tissues were compared in the ONCOMINE, UALCAN and HPA databases. ONCOMINE differential expression analysis revealed that the transcriptional levels of XPO1, CSE1L, XPOT, and XPO5 were upregulated in patients with LUAD and LUSC (Figure 1 and Table 1). The transcription level of XPO1 was significantly higher in patients with LUAD and LUSC in the 3 data sets. In the Hou Lung data set (27), XPO1 overexpression was detected in LUAD and LUSC compared with normal tissues, with fold changes of 1.375 (P=1.09e-09) and 2.828 (P=6.44e-09), respectively. However, in the TCGA Lung 2 data set, the XPO1 transcription level in LUAD and LUSC samples was 1.043 times (P=1.78e-12) and 1.179 times (P=3.11e-39) greater, respectively. Moreover, in the Stearman Lung data set, there was a 1.35-fold increase in XPO1 mRNA expression in LUAD tissues (P=1.95e-05) (28). The CSE1L transcriptional level in patients with LUAD and LUSC was also found to be elevated in the 3 data sets. In the Hou Lung data set (27), CSE1L was overexpressed in LUAD and LUSC compared with the normal samples, with a fold change of 1.487 (P=0.01e-08) and 2.51 (P=1.54e-17), respectively. In the Bhattacharjee Lung data set (29), CSE1L was overexpressed in LUSC with fold changes of 7.468 (P=0.67e-06), and the transcription level of CSE1L in LUAD was slightly higher than that in normal lung tissue, but the P value did not exceed 0.05. Furthermore, CSE1L was also overexpressed in LUAD and LUSC in the Garber Lung data set, with fold changes of 2.29 (P=0.001) and 3.497 (P=5.78e-05) (30). Similarly, the XPOT transcription level in patients with LUAD and LUSC was also elevated in the 3 data sets. In the Hou Lung data set (27) and Garber Lung data set (30), XPOT was significantly overexpressed in LUAD with fold changes of 1.572 (P=3.86E-13) and 1.768 (P=0.002), respectively.
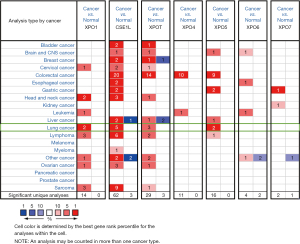
Table 1
| Gene | Type of lung cancer versus normal lung tissue | Fold change | P value | t-test | Source and/or reference |
|---|---|---|---|---|---|
| XPO1 | LUAD | 1.375 | 1.09E-09 | 6.704 | Hou (27) |
| LUSC | 2.828 | 6.44E-09 | 7.792 | Hou (27) | |
| LUSC | 1.179 | 3.11E-39 | 14.842 | TCGA | |
| LUAD | 1.043 | 1.78E-12 | 7.296 | TCGA | |
| LUAD | 1.35 | 1.95E-05 | 4.689 | Stearman (28) | |
| CSE1L | LUSC | 2.51 | 1.54E-17 | 13.493 | Hou (27) |
| LUAD | 1.487 | 2.01E-08 | 6.38 | Hou (27) | |
| LUSC | 7.468 | 7.67E-06 | 4.994 | Bhattacharjee (29) | |
| LUAD | 1.566 | 0.065 | 1.588 | Bhattacharjee (29) | |
| LUSC | 3.497 | 5.78E-05 | 5.227 | Garber (30) | |
| LUAD | 2.29 | 0.001 | 4.431 | Garber (30) | |
| XPOT | LUAD | 1.075 | 0.382 | 0.304 | Bhattacharjee (29) |
| LUSC | 4.078 | 9.25E-05 | 4.166 | Bhattacharjee (29) | |
| LUSC | 2.527 | 5.04E-13 | 11.371 | Hou (27) | |
| LUAD | 1.572 | 3.86E-11 | 7.771 | Hou (27) | |
| LUAD | 1.768 | 0.002 | 3.571 | Garber (30) | |
| XPO5 | LUSC | 2.044 | 1.72E-11 | 9.733 | Hou (27) |
| LUAD | 1.684 | 1.63E-10 | 7.593 | Hou (27) | |
| LUSC | 2.161 | 0.001 | 3.977 | Garber (30) | |
| LUAD | 2.126 | 0.002 | 4.505 | Garber (30) |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
In the Bhattacharjee Lung data set (29), the transcription level of XPOT in LUSC had an increased fold change of 4.078 (P=9.25e-05), but the same change was not found in LUAD samples (P=0.382), compared with normal tissues. A similar pattern for XPO5 was found in the Hou Lung data set (27). XPO5 was significantly upregulated in LUAD and LUSC with fold changes of 1.684 (P=1.63e-10) and 2.044 (P=1.72e-11) respectively. Similar results were apparent in the Garber Lung data set (30).
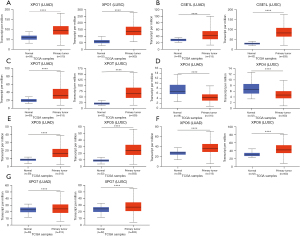
Next, the transcriptional expression of exportins members between LUAD, LUSC and normal tissues was confirmed by using the UALCAN data set (Figure 2). The results indicated that the transcription levels of XPO1, CSE1L, XPOT, XPO5, XPO6, and XPO7 in LUAD and LUSC tissues were significantly higher than those in normal lung tissues, while the transcription level of XPO4 in normal lung tissues was higher than that in LUAD and LUSC tissues.
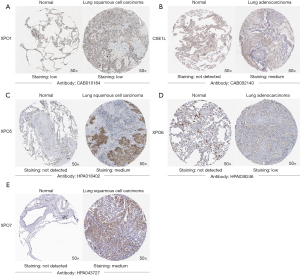
After analyzing the transcription levels of exportins in LUAD and LUSC, we confirmed the protein expression levels of exportins in patients with LUAD and LUSC and normal lung tissues using the HPA database (Figure 3). It should be noted that XPO1 protein was expressed in both normal lung tissues and lung cancer tissues (Figure 3A). CSE1L, XPO5, XPO6, and XPO7 were not expressed in normal lung tissues, but had low and medium expression in lung cancer tissues (Figure 3B-3E). Moreover, there was no protein expression of XPOT and XPO4 in normal or lung cancer tissues. In summary, the results of the above 3 databases revealed that the transcriptional and translational expression levels of the XPO1, CSE1L, XPO5, XPO6, and XPO7 genes were increased in patients with LUAD and LUSC.
Prognostic characteristics of exportin members in patients with LUAD and LUSC
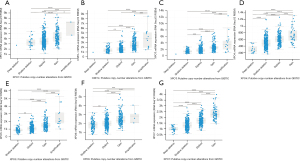
In order to evaluate the clinical significance of exportins, the publicly accessible Kaplan-Meier plotter tool was used to determine the correlation between exportin members’ transcriptional levels and the survival of patients with LUAD and LUSC. The main parameters of survival analysis included overall survival (OS) and progression-free survival (PFS). According to the Kaplan-Meier survival curves shown in Figure 4 and Table 2, OS was negatively correlated with the transcriptional levels of CSE1L and XPOT/4/5/6/7 but positively correlated with XPO1 (Figure 4A-4G). Moreover, the transcriptional levels of XPO4/6/7 in LUAD and LUSC were negatively correlated with PFS, while the transcriptional levels of XPO1 and XPOT were positively correlated with PFS, but there was no significant correlation between the transcriptional levels of CSE1L and XPO5 with PFS (Figure 4A-4G). In summary, according to the transcriptional and protein expression levels of exportins in the above databases and the results of the Kaplan-Meier plotter tool, the elevated transcriptional levels of CSE1L and XPO5/6/7 were associated with a worse prognosis in patients with LUAD and LUSC. In contrast, increased XPO1 transcription levels were associated with a better prognosis in patients with LUAD and LUSC. Therefore, CSE1L and XPO1/5/6/7 may be useful biomarkers for predicting the survival rate of patients with LUAD and LUSC.
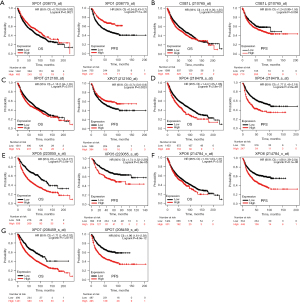
Table 2
| Exportin | Histology | OS | PFS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | HR | 95% CI | P value | Cases | HR | 95% CI | P value | |||
| XPO1 | Overall | 1,925 | 0.79 | 0.68–0.92 | 0.0021 | 982 | 0.54 | 0.43–0.71 | 1.00E-06 | |
| 208775_at | LUAD | 719 | 0.27 | 0.19–0.38 | 1.50E–15 | 461 | 0.55 | 0.4–0.75 | 0.00017 | |
| LUSC | 524 | 1.07 | 0.84–1.35 | 0.6 | 141 | 1.89 | 1.12–3.2 | 0.016 | ||
| CSE1L | Overall | 1,925 | 1.19 | 1.05–1.35 | 0.0074 | 982 | 1.24 | 0.99–1.55 | 0.058 | |
| 210765_at | LUAD | 719 | 1.35 | 1.04–1.73 | 0.021 | 461 | 0.7 | 0.51–0.96 | 0.026 | |
| LUSC | 524 | 0.58 | 0.66–1.08 | 0.19 | 141 | 1.67 | 1–2.78 | 0.048 | ||
| XPOT | Overall | 1,925 | 1.18 | 1.04–1.35 | 0.014 | 982 | 0.74 | 0.6–0.9 | 0.0025 | |
| 212160_at | LUAD | 719 | 0.58 | 0.46–0.74 | 4.60E–06 | 461 | 0.69 | 0.5–0.95 | 0.022 | |
| LUSC | 524 | 0.89 | 0.67–1.18 | 0.42 | 141 | 1.79 | 1.05–3.05 | 0.031 | ||
| XPO4 | Overall | 1,925 | 1.44 | 1.25–1.68 | 2.90E–07 | 982 | 1.77 | 1.45–2.18 | 2.80E-08 | |
| 218479_s_at | LUAD | 719 | 0.86 | 0.68–1.08 | 0.19 | 461 | 1.59 | 1.15–2.2 | 0.0051 | |
| LUSC | 524 | 1.34 | 1.04–1.72 | 0.025 | 141 | 2.42 | 1.34–4.36 | 0.0025 | ||
| XPO5 | Overall | 1,144 | 1.9 | 1.6–2.27 | 3.80E–13 | 596 | 1.74 | 1.32–2.28 | 5.90E-05 | |
| 223055_s_at | LUAD | 672 | 1.76 | 1.38–2.24 | 3.40E–06 | 443 | 1.9 | 1.38–2.63 | 7.40E-05 | |
| LUSC | 271 | 1.59 | 1.09–2.32 | 0.014 | 141 | 1.61 | 0.97–2.69 | 0.065 | ||
| XPO6 | Overall | 1,925 | 1.19 | 1.03–1.37 | 0.02 | 982 | 1.68 | 1.39–2.04 | 8.10E-08 | |
| 214784_x_at | LUAD | 719 | 1.47 | 1.12–1.93 | 0.0053 | 461 | 1.66 | 1.22–2.28 | 0.0013 | |
| LUSC | 524 | 0.82 | 0.63–1.07 | 0.14 | 141 | 1.47 | 0.84–2.56 | 0.17 | ||
| XPO7 | Overall | 1925 | 1.71 | 1.45–2.02 | 1.20E–10 | 982 | 1.96 | 1.61–2.38 | 8.90E-12 | |
| 208459_s_at | LUAD | 719 | 2.24 | 1.76–2.84 | 1.10E–11 | 461 | 2.04 | 1.47–2.83 | 1.30E-05 | |
| LUSC | 524 | 0.84 | 0.64–1.1 | 0.21 | 141 | 1.54 | 0.9–2.63 | 0.11 | ||
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval.
The genetic alteration and mutation information of exportins
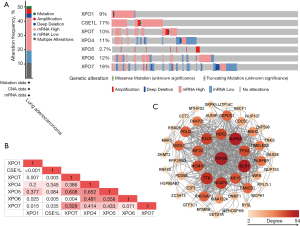
The LUAD (TCGA, Firehose legacy) module was used in cBioPortal online tool to analyze the mutations of exportin members. As shown in Figure 5A, 261 of 517 patients had gene mutations (mutation rate 50.48%), of which CSE1L and XPO7 were the genes with the largest mutations, with mutation rates of 17% and 18%, respectively. Furthermore, the mutation rates of the XPO1, XPOT, XPO4, XPO5, and XPO6 genes were 9%, 10%, 1%, 11%, 2.7%, and 12% in the LUAD samples, respectively. Gene mutations in exportins members included mRNA upregulation (120 cases, 23.21%), multiple alterations (56 cases, 10.83%), mRNA downregulation (44 cases, 8.51%), genetic amplification (18 cases, 3.48%), deep deletion (14 case, 2.71%), and mutation (9 cases, 1.74%). Among them, the highest proportion of mutation was mRNA upregulation, especially in XPO1, CSE1L, XPOT, and XPO6. However, the overexpression of mRNA was not detected in XPO5, but it had high frequency of genetic amplification of 2.7%. we also figured the correlations between exportins family by analyzing their mRNA expression (RNA Seq V2 RSEM) via the cBioPortal online tool for LUAD (TCGA, Firehose Legacy). The results showed that XPO1 was significantly and positively correlated with CSE1L, XPOT, XPO4, XPO6, and XPO7; CSE1L was positively correlated with XPOT, XPO5, XPO6 and XPO7; and XPOT was significantly and positively correlated with XPO6 (Figure 5B). In addition, the “mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM)” option in the genome map was selected to analyze the correlation between exportin members through the “mutual exclusivity” module. The results (Figure 5C) indicated that there was a correlation between the transcriptional levels of XPO1, CSE1L, XPOT, and XPO6, and that the transcriptional levels of XPO1 and CSE1L were also correlated with XPO7. Furthermore, the relationship between the mRNA expression and copy-number alterations of exportins members was analyzed, and it was found that the mRNA expression of exportins members was positively correlated with copy-number alterations (Figure 6).
GO and KEGG enrichment analysis of exportins and their coexpression genes in patients with LUAD and LUSC
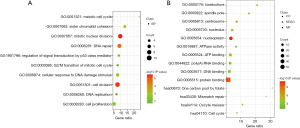
Subsequently, the “Coexpression” module of cBioPortal was used to list the top ten co-expression genes with the largest Pearson’s correlation coefficient among the seven exportins, and a total of 70 genes were listed (Table S1). After the deletion of duplicate genes, a total of 69 genes were selected. Following this, the STRING database was used to construct the PPI network of the 69 coexpression genes that were significantly related to exportins mutations, with the module analysis being conducted using Cytoscape software. As shown in Figure 5C, the top 11 genes significantly associated with the exportins mutations were TOP2A, AURKA, BUB1, EXO1, TTK, MCM10, NCAPG, KIF2C, NEK2, KIF15, and TPX2. Next, the biological functions of the 69 coexpression genes significantly associated with exportins mutations were further analyzed through GO enrichment analysis and KEGG pathway enrichment in DAVID. As shown in Figure 7 and Table S2, it was found that biological processes of mitotic nuclear division (GO:0007067), cell population proliferation (GO:0008283), regulation of signal transduction by p53 class mediator (GO:1901796), G2/M transition of mitotic cell cycle (GO:0000086), and G2/M transition of mitotic cell cycle (GO:0000086) were regulated by exportins mutations in NSCLC (Figure 7A). Moreover, the cellular components of nucleoplasm (GO:0005654), centrosome (GO:0005813), kinetochore (GO:0000776), nucleolus (GO:0005730), and spindle pole (GO:0000922) were significantly associated with the exportin alterations (Figure 7B). Furthermore, exportins mutations also prominently affected the molecular functions of protein binding (GO:0005515), adenosine triphosphate (ATP) binding (GO:0005524), poly (A) RNA binding (GO:0044822), DNA binding (GO:0003677 and ATP hydrolysis activity (GO:0016887) (Figure 7B). In KEGG analysis, oocyte meiosis was closely related to the function of exportins in NSCLC, and cell cycle pathway was also found to be related to exportins, although nonsignificantly (P>0.05) (Table S2). The above results suggest that the functions of exportins mutations and their coexpressed genes may be associated with cell proliferation, cell division, regulation of signal transduction by p53 class mediator, and cell cycle, among other processes.
The relationship between exportins expression levels and immune infiltration levels in LUAD and LUSC
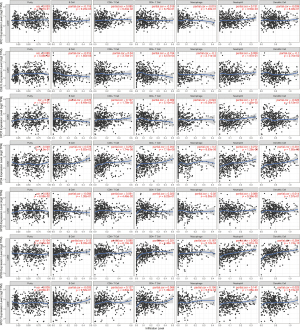
Immunity is closely related to the occurrence and development of tumors. Therefore, the relationship between the transcriptional levels of exportins and the levels of immune infiltration in LUAD and LUSC was evaluated using the TIMER online analysis tool. It was found that exportins were associated with immune cell infiltration. As shown in Figure 8, XPO1 was negatively correlated with B cells infiltration and positively correlated with neutrophils infiltration. CSE1L was negatively correlated with B cells infiltration, CD4+ T cells infiltration, macrophages infiltration, neutrophils infiltration, and dendritic cells infiltration. Additionally, XPO4 and XPO6 were positively correlated with CD4+ T cells infiltration, macrophages infiltration, neutrophils infiltration, and dendritic cells infiltration; XPO4 was positively correlated with CD8+ T cells infiltration; XPO6 was also positively correlated with B cells infiltration; and XPO7 was positively correlated with CD8+ T cells infiltration, macrophages infiltration, and neutrophils infiltration. However, it was discovered that XPO5 was only positively correlated with CD8+ T cells infiltration. These studies indicated that the expression levels of exportins were correlated with the levels of immune infiltration in LUAD and LUSC.
Analysis of transcription factors and miRNA targets of exportins in patients with LUAD and LUSC
Finally, the transcription factor targets (TFs) and mRNA targets of differentially expressed exportins from the LinkedOmics database were analyzed. The first 3 enriched miRNA targets of each exportins are shown in Table 3. It was found that miR-433 (ATCATGA) was the common target of XPO1 and CSE1L, and miR-26A, miR-26B (TACTTGA) were the common targets of XPOT and XPO4. Similarly, the common targets of XPO4 and XPO7 were miR-181A, miR-181B, miR-181C, miR-181D (TGAATGT), while miR-452 (GAGACTG) was the common target of XPO5 and XPO7. Finally, it was found that E2F1 appears to be the key TF regulated by exportins (Table 4).
Table 3
| Gene | Enriched miRNA target | Leading edge, n | P value |
|---|---|---|---|
| XPO1 | ATCATGA, MIR-433 | 45 | <0.0001 |
| ATTCTTT, MIR-186 | 75 | <0.0001 | |
| CATGTAA, MIR-496 | 47 | <0.0001 | |
| CSE1L | ATAACCT, MIR-154 | 20 | <0.0001 |
| GAGCCTG, MIR-484 | 37 | <0.0001 | |
| ATCATGA, MIR-433 | 35 | <0.0001 | |
| XPOT | AGTCTTA, MIR-499 | 26 | <0.0001 |
| TACTTGA, MIR-26A, MIR-26B | 69 | <0.0001 | |
| TAGCTTT, MIR-9 | 63 | 0.003 | |
| XPO4 | TGAATGT, MIR-181A, MIR-181B, MIR-181C, MIR-181D | 156 | <0.0001 |
| CAGTGTT, MIR-141, MIR-200A | 112 | <0.0001 | |
| TACTTGA, MIR-26A, MIR-26B | 109 | <0.0001 | |
| XPO5 | GTTTGTT, MIR-495 | 82 | <0.0001 |
| GTGGTGA, MIR-197 | 28 | <0.0001 | |
| GAGACTG, MIR-452 | 42 | <0.0001 | |
| XPO6 | CAGCAGG, MIR-370 | 43 | <0.0001 |
| AGGGCCA, MIR-328 | 25 | <0.0001 | |
| CACTGCC, MIR-34A, MIR-34C, MIR-449 | 94 | <0.0001 | |
| XPO7 | CTTGTAT, MIR-381 | 75 | <0.0001 |
| TGAATGT, MIR-181A, MIR-181B, MIR-181C, MIR-181D | 180 | <0.0001 | |
| GAGACTG, MIR-452 | 27 | <0.0001 |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Table 4
| Gene | Enriched transcription factor target | Leading edge, n | P value |
|---|---|---|---|
| XPO1 | V$E2F_Q4 | 86 | <0.0001 |
| V$E2F_Q6 | 86 | <0.0001 | |
| V$E2F1_Q6 | 84 | <0.0001 | |
| CSE1L | V$E2F_Q6 | 99 | <0.0001 |
| V$E2F1DP1_01 | 93 | <0.0001 | |
| V$E2F1DP2_01 | 93 | <0.0001 | |
| XPOT | V$E2F_Q4 | 82 | <0.0001 |
| V$E2F_Q6 | 82 | <0.0001 | |
| V$E2F1_Q6 | 85 | <0.0001 | |
| XPO4 | V$NFMUE1_Q6 | 66 | <0.0001 |
| V$HTF_01 | 17 | <0.0001 | |
| V$STAT5A_04 | 71 | <0.0001 | |
| XPO5 | V$E2F_Q4 | 107 | <0.0001 |
| V$E2F_Q6 | 108 | <0.0001 | |
| V$E2F1_Q6 | 108 | <0.0001 | |
| XPO6 | V$E2F_Q6 | 86 | <0.0001 |
| V$E2F_Q4 | 86 | <0.0001 | |
| V$E2F1DP1_01 | 86 | <0.0001 | |
| XPO7 | V$HTF_01 | 17 | <0.0001 |
| V$RREB1_01 | 45 | <0.0001 | |
| V$E2F_Q6 | 62 | <0.0001 |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Discussion
Exportin members have been partially shown to be associated with a variety of different tumors, such as liver cancer (9), breast cancer (10,31), gastric cancer (11), and lung cancer (12). One study found that the stable overexpression of CRM1 in human bronchial epithelial cells leads to malignant cellular transformation (14). XPO1 knockout enhanced the sensitivity of SCLC cells to chemotherapy, and XPO1 inhibition showed synergistic effect with first-line and second-line chemotherapy. Selinur is a small molecule XPO1 inhibitor, which can significantly inhibit tumor growth in patients with SCLC in combination with cisplatin or ibrutinib (32). In our study, the expression level, genetic variation, immune infiltration, biological function of different exportins, and their relationship with the prognosis of patients with LUAD and LUSC were comprehensively analyzed by bioinformatics for the first time.
XPO1 was identified as a gene required to modify chromosome structures or regulate the maintenance of high-order chromosome structures in Schizosaccharomyces pombe, so it was originally named CRM1 (chromosome region maintenance 1). Subsequently, it was found that it had homology with importin beta-like transporter and could specifically interact with NES motif and Ran GTPase to mediate the nuclear export of protein and mRNA in Saccharomyces cerevisiae, and it was finally renamed XPO1 (export 1) (33). As a well-defined nuclear export protein, XPO1 is responsible for exporting a variety of proteins and RNAs [including ribosomal RNA, small nuclear RNA, mRNA, microRNA, and transfer RNA (tRNA)] (34), and plays an important role in regulating mitosis and chromosome structure (35), which indicates that many carcinogenic mechanisms may involve XPO1. In eukaryotic cells, XPO1 has been considered to be an important exporter of most tumor suppressor proteins, including p53, p21, PI3K/Akt, p27, and BRCA1/2, all of which are important targets for tumorigenesis (36). XPO1 has been shown to be highly expressed in many types of malignant tumors and associated with poor prognosis, including ovarian cancer (37), prostate cancer (38), osteosarcoma (39), leukemia (40), multiple myeloma (41,42), and glioma (43). One study demonstrated CRM1 overexpression in lung cancer, with CRM1 being frequently overexpressed in NSCLC, especially adenocarcinoma and squamous cell carcinoma, which may be caused by the synergistic effect of CRM1 overexpression and p53 phosphorylation in cell malignant transformation. Meanwhile, inhibiting CRM1 can improve the efficacy of cisplatin in the treatment of lung cancer (14). In our study, we found through database analysis that the transcriptional level of XPO1 in patients with LUAD and LUSC was higher than that in normal tissues. Nagasaka et al. (44) retrospectively analyzed de-identified pathological and molecular information from 18,218 NSCLC samples to describe the prevalence of XPO1 mutations and amplifications in NSCLC. Their study found that presence of XPO1 pathogenic mutations was associated with a poor OS in both the entire NSCLC cohort and the adenocarcinoma subgroup. However, Li et al. (45) divided patients with NSCLC into a pure XPO1 mutant, wild-type, and XPO1-STK11/KEAP1 mutant groups and found that patients with pure XPO1 mutations had longer survival. So, in order to study the relationship between XPO1 and the prognosis of lung cancer, we used Kaplan-Meier plotter to analyze the findings that in patients with NSCLC, increased XPO1 transcript level was associated with better OS and PFS. We believe that this may be related to the XPO1 mutation, which renders NSCLC more sensitive to treatment and thus improves prognosis. Our study also confirmed that XPO1 has a high mutation rate in LUAD, with a mutation rate of 9%. This finding suggests that XPO1 may be a valuable prognostic marker for NSCLC, but further studies are still needed.
CSE1L, also known as human cellular apoptosis susceptibility and exportin 2 (XPO2), has been shown to be involved in the regulation of different cellular mechanisms, including mitotic spindle checkpoint, proliferation, and apoptosis (46). However, the role of CSE1L in lung cancer has rarely been studied. A previous study showed that ACOT11 can promote the proliferation, migration, and invasion of lung cancer cells by binding with CSE1L (47). CSE1L can regulate the MAPK pathway and promote the development of lung cancer by interacting with p65 (15). Another study reported that CSE1L silencing impaired the cell proliferation, invasion, and migration of lung cancer cells, and that the expression of STAT3 and MET protein was decreased; the apoptosis-related marker protein c-PARP was induced, which indicated that cell proliferation and antiapoptosis may be regulated by CSE1L via the MET–STAT3 pathway (48). In our study, ONCOMINE and UALCAN databases revealed that the transcriptional level of CSE1L in LUAD and LUSC was higher than that in normal tissues. HPA database also found that CSE1L was not expressed in normal lung tissue, but was moderately expressed in LUAD tissue. Additionally, Kaplan-Meier plotter analysis revealed that the high expression level of CSE1L was significantly linked with a worse OS but not with PFS in patients with NSCLC.
XPOT and XPO5 are Kap-β family of tRNA nuclear output receptors. XPOT is a specific nuclear export receptor of tRNA, and XPO5 mainly exports miRNA to the cytoplasm, and thus the role of XPO5 in tRNA export is considered to be secondary (49). Previous studies have reported the key roles of XPOT in the development and progression of various tumors, such as breast cancer (50) and human promyelocytic leukemia (51). The overexpression of XPOT was reported to lead to more aggressive and chromosomally unstable breast cancers (50), while the elevated expression of XPOT was associated with cell proliferation in human promyelocytic leukemia (51). In addition, Pan et al. (52) have found in recent years that knocking out XPOT through small interfering RNA can inhibit the proliferation and migration of neuroblastoma cells. However, the role of XPOT in lung cancer has not been previously explored. Regarding XPO5, Özdaş et al. (53) found that the mRNA and protein levels of XPO5 were upregulated in metastatic cells of head and neck squamous cell carcinoma (HNSCC), and silencing XPO5 resulted in reduced cell proliferation, delayed wound healing, and increased Caspase-3 enzyme activity in HNSCC cell lines. A previous report indicated that the miR-single-nucleotide polymorphism (SNP) of rs11077 in the miRNA processing gene XPO5 was associated with the recurrence of resected NSCLC, which suggests that the miR-SNP in the miRNA processing mechanism gene of XPO5 is involved in the prognosis of NSCLC (54). In our study, the ONCOMINE database analysis results in transcriptional levels of XPOT and XPO5 in NSCLC being significantly higher than those in normal lung tissue, and these results were corroborated by the UALCAN data set. Immunohistochemical staining demonstrated that XPO5 was moderately expressed in lung cancer tissues, but not in normal tissues. However, XPOT expression was not found in lung cancer or normal tissues. Furthermore, Kaplan-Meier plotter analysis indicated that the high expression of XPO5 was associated with worse OS. Therefore, we believe that XPO5 may be an important indicator for predicting the prognosis of patients with NSCLC.
XPO4 is a bidirectional nuclear transporter involved in the nuclear export of translation initiation factor EIF5A and transcriptional regulator Smad3 (55,56). Smad3, an effector of tumor growth factor (TGF)-β signal transduction, can promote or inhibit the growth of cancer cells (57). EIF-5A has been identified as the initiation gene of eukaryotic translation and to be encoded by 2 highly related genes (EIF5A1 and EIF5A2). Although the mechanism of EIF5A1 causing cancers has not been fully clarified, a study found that the EIF5A1 and EIF5A2 proteins are overexpressed in some human tumors (58). At present, there are no more reports on the relationship of XPO4 with any cancers. We found that the ONCOMINE data set did not record the expression of XPO4 transcription level in lung cancer and that the HPA data set did not contain the expression of XPO4 protein in lung cancer or normal tissues. Due to the lack of relevant studies on XPO4, its role in the occurrence and development of NSCLC cannot be confirmed.
Similar to XPO4, there are relatively few reports on XPO6/7 in NSCLC. The UALCAN data set in our study revealed that the transcriptional levels of XPO6 and XPO7 in LUAD and LUSC were higher than those in normal tissues. Immunohistochemical staining revealed that XPO6 and XPO7 were low and moderately expressed in lung cancer tissues but not in normal tissues. Survival analysis demonstrated that the high expressions of XPO6 and XPO7 were associated with worse OS and PFS in patients with LUAD and LUSC. Based on these results, we speculate that XPO6/7 may be a potential biomarker for patients with NSCLC, but this needs to examined in further study.
Furthermore, it was found that exportins had a high mutation rate (50.48%) in patients with LUAD and LUSC. The highest mutation was mRNA upregulation, and a co-occurring relationship was found between differentially expressed exportins, which indicated that these genes play a synergistic role in the occurrence and development of LUAD and LUSC. A network of coexpression genes significantly associated with exportins mutations was constructed, and then their functions were analyzed through GO enrichment analysis and KEGG pathway enrichment. Our studies have found that the functions of these proteins are mainly related to cell proliferation, cell division, regulation of signal transduction by p53 class mediator, cell cycle, and other processes. These functions are significantly related to the occurrence and development of tumors. These results provide clues for the rational development of multitarget and exportin-mediated targeted therapy.
Recent data have suggested that as an important determinant of prognosis and response to immunotherapy immune cell (59), infiltration may affect tumor progression and recurrence (60). Tumor-derived microbubbles (TMVs) are extracellular vesicles released from tumor cells, which are now understood to promote the communication between tumor and surrounding microenvironment. A research showed that the interaction between ARF6-GTP and XPO5 transported a pre-miRNA complex to sites of TMV biogenesis for inclusion as TMV cargo. This indicates that XPO5 is related to tumor microenvironment (61). Our study conducted an in-depth investigation of the infiltration of 6 immune cells (B cells, CD8 + T cells, CD4 + T cells, macrophages, neutrophils, and dendritic cells) related to exportins. We found that the expression levels of exportins were found to be associated with the levels of immune infiltration in LUAD and LUSC, which can reflect the immune status indicators of patients with LUAD and LUSC.
We also sought to characterize the transcription miRNA targets of the differentially expressed exportins and found that several miRNAs (miR-433, miR-26, miR-181 and miR-452) were associated with differentially expressed exportins. These small noncoding RNAs mainly promote tumor growth, invasion, angiogenesis, and immune escape by controlling the expression of their target mRNAs. Moreover, tumor miRNA distribution can define related subtypes, patient survival, and treatment response, and it plays an important role in cancer biology (62). It was found that decreased miR433 expression was strongly associated with the tumor-node-metastasis stage and lymph node metastasis of patients with NSCLC (63). miR‑433 reduces cell proliferation and invasive phenotype in patients with NSCLC by mediating Smad2 and Id-1 (64). Furthermore, miR-181 and miR-452 also play an important role in epithelial-mesenchymal transition (EMT), proliferation, invasion, and metastasis of lung cancer cells (65,66). Our analysis suggested that differentially expressed exportins may play a role through these miRNAs to regulate the occurrence and development of LUAD and LUSC.
By mining the TF targets of differentially expressed exportins, it was found that E2F1 may the key TF regulated by exportins. E2F1 is one of the key links in the cell cycle regulation network (67) and is a central player in cell cycle progression, DNA-damage response, and apoptosis (68). A study proven that E2F1 promotes EMT by regulating ZEB2 in SCLC (69). In SCLC cells, ILF2 interacts with E2F1 and regulates the transcriptional activity of E2F1, exerting a carcinogenic effect (70). Our analysis revealed that E2F1 is a promising regulatory target of exportins, as differentially expressed exportins may regulate cell genesis and development by interacting with E2F1 in LUAD and LUSC.
Conclusions
This study is the first to systematically demonstrated the association between exportins and LUAD and LUSC. We further showed that the transcriptional and protein expression levels of CSE1L and XPO1/5/6/7 were increased in patients with LUAD and LUSC, and the increased transcriptional levels of CSE1L and XPO5/6/7 were associated with worse prognosis, with an increased transcriptional level of XPO1 suggesting a better prognosis. These results point to CSE1L and XPO1/5/6/7 as potential prognostic biomarkers for the survival of patients with LUAD and LUSC. Furthermore, it was found that the expression of exportins was significantly correlated with the infiltration of a variety of immune cells. Differentially expressed exportins regulate the occurrence and development of LUAD and LUSC by involving a variety of miRNAs along with transcription factor E2F1. Our study may provide novel insights into the selection of prognostic biomarkers of exportins in LUAD and LUSC. However, our research still has some limitations. Since all the data in our study are from the public databases, there is no cellular function researches or animal experiments in vivo and in vitro of exportins, and then we will start to carry out relevant experimental researches to promote the clinical application of exportins as potential biomarkers of LUAD and LUSC.
Acknowledgments
Funding: This work was supported by Guangxi Medical and Health Key Cultivation Discipline Construction Project; Scientific Research Project of the Affiliated Hospital of Youjiang Medical College for Nationalities in 2021 (No. y20212607).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-228/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-228/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-228/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Chen Z, Chen X, Lei T, et al. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther 2020;28:1479-93. [Crossref] [PubMed]
- Muqbil I, Kauffman M, Shacham S, et al. Understanding XPO1 target networks using systems biology and mathematical modeling. Curr Pharm Des 2014;20:56-65. [Crossref] [PubMed]
- Cautain B, Hill R, de Pedro N, et al. Components and regulation of nuclear transport processes. FEBS J 2015;282:445-62. [Crossref] [PubMed]
- Xu D, Grishin NV, Chook YM. NESdb: a database of NES-containing CRM1 cargoes. Mol Biol Cell 2012;23:3673-6. [Crossref] [PubMed]
- Chook YM, Süel KE. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta 2011;1813:1593-606. [Crossref] [PubMed]
- Soniat M, Chook YM. Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J 2015;468:353-62. [Crossref] [PubMed]
- Lin J, Hou Y, Huang S, et al. Exportin-T promotes tumor proliferation and invasion in hepatocellular carcinoma. Mol Carcinog 2019;58:293-304. [Crossref] [PubMed]
- Tai CJ, Shen SC, Lee WR, et al. Increased cellular apoptosis susceptibility (CSE1L/CAS) protein expression promotes protrusion extension and enhances migration of MCF-7 breast cancer cells. Exp Cell Res 2010;316:2969-81. [Crossref] [PubMed]
- Sun YQ, Xie JW, Xie HT, et al. Expression of CRM1 and CDK5 shows high prognostic accuracy for gastric cancer. World J Gastroenterol 2017;23:2012-22. [Crossref] [PubMed]
- Geng JQ, Wang XC, Li LF, et al. MicroRNA-related single-nucleotide polymorphism of XPO5 is strongly correlated with the prognosis and chemotherapy response in advanced non-small-cell lung cancer patients. Tumour Biol 2016;37:2257-65. [Crossref] [PubMed]
- Hao J, Chiang YT, Gout PW, et al. Elevated XPO6 expression as a potential prognostic biomarker for prostate cancer recurrence. Front Biosci (Schol Ed) 2016;8:44-55. [Crossref] [PubMed]
- Gao W, Lu C, Chen L, et al. Overexpression of CRM1: A Characteristic Feature in a Transformed Phenotype of Lung Carcinogenesis and a Molecular Target for Lung Cancer Adjuvant Therapy. J Thorac Oncol 2015;10:815-25. [Crossref] [PubMed]
- Lin HC, Li J, Cheng DD, et al. Nuclear export protein CSE1L interacts with P65 and promotes NSCLC growth via NF-κB/MAPK pathway. Mol Ther Oncolytics 2021;21:23-36. [Crossref] [PubMed]
- Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol 2011;671:3-34. [Crossref] [PubMed]
- Guo P, Xu T, Jiang Y, et al. Identification of potential key molecular biomarkers in lung adenocarcinoma by bioinformatics analysis. Transl Cancer Res 2022;11:227-41. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010;5:e10312. [Crossref] [PubMed]
- Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol 2005;167:1763-75. [Crossref] [PubMed]
- Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 2001;98:13790-5. [Crossref] [PubMed]
- Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001;98:13784-9. [Crossref] [PubMed]
- Zhu C, Kim SJ, Mooradian A, et al. Cancer-associated exportin-6 upregulation inhibits the transcriptionally repressive and anticancer effects of nuclear profilin-1. Cell Rep 2021;34:108749. [Crossref] [PubMed]
- Quintanal-Villalonga A, Taniguchi H, Hao Y, et al. Inhibition of XPO1 Sensitizes Small Cell Lung Cancer to First- and Second-Line Chemotherapy. Cancer Res 2022;82:472-83. [Crossref] [PubMed]
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol 1989;108:1195-207. [Crossref] [PubMed]
- Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 2007;17:193-201. [Crossref] [PubMed]
- Ho CY, Wong CH, Li HY. Perturbation of the chromosomal binding of RCC1, Mad2 and survivin causes spindle assembly defects and mitotic catastrophe. J Cell Biochem 2008;105:835-46. [Crossref] [PubMed]
- Gravina GL, Senapedis W, McCauley D, et al. Nucleo-cytoplasmic transport as a therapeutic target of cancer. J Hematol Oncol 2014;7:85. [Crossref] [PubMed]
- Landes JR, Moore SA, Bartley BR, et al. The efficacy of selinexor (KPT-330), an XPO1 inhibitor, on non-hematologic cancers: a comprehensive review. J Cancer Res Clin Oncol 2023;149:2139-55. [Crossref] [PubMed]
- Gravina GL, Mancini A, Sanita P, et al. Erratum to: KPT-330, a potent and selective exportin-1 (XPO-1) inhibitor, shows antitumor effects modulating the expression of cyclin D1 and survivin in prostate cancer models. BMC Cancer 2016;16:8. [Crossref] [PubMed]
- Jiang Y, Hou J, Zhang X, et al. Circ-XPO1 upregulates XPO1 expression by sponging multiple miRNAs to facilitate osteosarcoma cell progression. Exp Mol Pathol 2020;117:104553. [Crossref] [PubMed]
- Lin KH, Rutter JC, Xie A, et al. P2RY2-AKT activation is a therapeutically actionable consequence of XPO1 inhibition in acute myeloid leukemia. Nat Cancer 2022;3:837-51. [Crossref] [PubMed]
- Tai YT, Landesman Y, Acharya C, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia 2014;28:155-65. [Crossref] [PubMed]
- Misund K, Hofste Op Bruinink D, Coward E, et al. Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence. Leukemia 2022;36:1887-97. [Crossref] [PubMed]
- Liu X, Chong Y, Tu Y, et al. CRM1/XPO1 is associated with clinical outcome in glioma and represents a therapeutic target by perturbing multiple core pathways. J Hematol Oncol 2016;9:108. [Crossref] [PubMed]
- Nagasaka M, Asad MFB, Al Hallak MN, et al. Impact of XPO1 mutations on survival outcomes in metastatic non-small cell lung cancer (NSCLC). Lung Cancer 2021;160:92-8. [Crossref] [PubMed]
- Li X, Zou B, Wang S, et al. XPO1-mutant NSCLC without STK11/KEAP1 mutations may predict better survival to immunotherapy. J Transl Med 2021;19:421. [Crossref] [PubMed]
- Zhu JH, Hong DF, Song YM, et al. Suppression of cellular apoptosis susceptibility (CSE1L) inhibits proliferation and induces apoptosis in colorectal cancer cells. Asian Pac J Cancer Prev 2013;14:1017-21. [Crossref] [PubMed]
- Liang C, Wang X, Zhang Z, et al. ACOT11 promotes cell proliferation, migration and invasion in lung adenocarcinoma. Transl Lung Cancer Res 2020;9:1885-903. [Crossref] [PubMed]
- Liu W, Zhou Z, Li Y, et al. CSE1L silencing impairs tumor progression via MET/STAT3/PD-L1 signaling in lung cancer. Am J Cancer Res 2021;11:4380-93. [PubMed]
- Okamura M, Inose H, Masuda S. RNA Export through the NPC in Eukaryotes. Genes (Basel) 2015;6:124-49. [Crossref] [PubMed]
- Vaidyanathan S, Thangavelu PU, Duijf PH. Overexpression of Ran GTPase Components Regulating Nuclear Export, but not Mitotic Spindle Assembly, Marks Chromosome Instability and Poor Prognosis in Breast Cancer. Target Oncol 2016;11:677-86. [Crossref] [PubMed]
- Suzuki T, Koyama Y, Hayakawa S, et al. 1,25-Dihydroxyvitamin D3 suppresses exportin expression in human promyelocytic leukemia HL-60 cells. Biomed Res 2006;27:89-92. [Crossref] [PubMed]
- Pan LJ, Chen JL, Wu ZX, et al. Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma. Technol Cancer Res Treat 2021;20:15330338211039132. [Crossref] [PubMed]
- Özdaş S, Canatar İ, Özdaş T. Effects of Knockdown of XPO5 by siRNA on the Biological Behavior of Head and Neck Cancer Cells. Laryngoscope 2022;132:569-77. [Crossref] [PubMed]
- Ding C, Li C, Wang H, et al. A miR-SNP of the XPO5 gene is associated with advanced non-small-cell lung cancer. Onco Targets Ther 2013;6:877-81. [PubMed]
- Lipowsky G, Bischoff FR, Schwarzmaier P, et al. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J 2000;19:4362-71. [Crossref] [PubMed]
- Kurisaki A, Kurisaki K, Kowanetz M, et al. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol 2006;26:1318-32. [Crossref] [PubMed]
- Teufel A, Staib F, Kanzler S, et al. Genetics of hepatocellular carcinoma. World J Gastroenterol 2007;13:2271-82. [Crossref] [PubMed]
- Clement PM, Johansson HE, Wolff EC, et al. Differential expression of eIF5A-1 and eIF5A-2 in human cancer cells. FEBS J 2006;273:1102-14. [Crossref] [PubMed]
- Liu X, Wu S, Yang Y, et al. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed Pharmacother 2017;95:55-61. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Clancy JW, Zhang Y, Sheehan C, et al. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat Cell Biol 2019;21:856-66. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Liu N, Liu Z, Zhang W, et al. MicroRNA 433 reduces cell proliferation and invasion in non small cell lung cancer via directly targeting E2F transcription factor 3. Mol Med Rep 2018;18:1155-64. [Crossref] [PubMed]
- Li J, Chen M, Yu B. miR-433 suppresses tumor progression via Smad2 in non-small cell lung cancer. Pathol Res Pract 2019;215:152591. [Crossref] [PubMed]
- He Z, Xia Y, Pan C, et al. Up-Regulation of MiR-452 Inhibits Metastasis of Non-Small Cell Lung Cancer by Regulating BMI1. Cell Physiol Biochem 2015;37:387-98. [Crossref] [PubMed]
- Huang P, Ye B, Yang Y, et al. MicroRNA-181 functions as a tumor suppressor in non-small cell lung cancer (NSCLC) by targeting Bcl-2. Tumour Biol 2015;36:3381-7. [Crossref] [PubMed]
- Polager S, Ginsberg D. E2F - at the crossroads of life and death. Trends Cell Biol 2008;18:528-35. [Crossref] [PubMed]
- Denechaud PD, Fajas L, Giralt A. E2F1, a Novel Regulator of Metabolism. Front Endocrinol (Lausanne) 2017;8:311. [Crossref] [PubMed]
- Wang T, Chen X, Qiao W, et al. Transcription factor E2F1 promotes EMT by regulating ZEB2 in small cell lung cancer. BMC Cancer 2017;17:719. [Crossref] [PubMed]
- Zhao M, Liu Y, Chang J, et al. ILF2 cooperates with E2F1 to maintain mitochondrial homeostasis and promote small cell lung cancer progression. Cancer Biol Med 2019;16:771-83. [Crossref] [PubMed]
(English Language Editor: J. Gray)


