Identification of autophagy-related genes and immune cell infiltration characteristics in sepsis via bioinformatic analysis
Highlight box
Key findings
• Autophagy-related genes may serve as biomarkers and may influence the development of sepsis by regulating immune function.
What is known and what is new?
• Autophagy is a key host defense mechanism against pathogens, plays a vital part in the induction and regulation of innate immune cell inflammatory responses, and influences the development of sepsis.
• The expression of ARGs may lead to the immune-activated microenvironment in the early stage of sepsis and may contribute to the diagnosis of sepsis.
What is the implication, and what should change now?
• Autophagy-related genes may influence the development of sepsis and have a vital impact on sepsis immune regulation.
Introduction
Sepsis is a disease caused by a dysregulated host response to infection that leads to severe organ dysfunction (1). Due to the high morbidity and mortality of sepsis, it is of great value to improve the prevention, recognition, and treatment of sepsis (2). In 2017, there were an estimated 48.9 million sepsis cases and 11 million sepsis-related deaths worldwide according to a study by the Institute for Health Metrics and Evaluation (IHME) on the global burden of sepsis (3). The pathogenic mechanism of the development of sepsis is immune dysfunction, including early immune system overactivation and late immune suppression (4). In the early stage of sepsis, immune cells such as macrophages secrete a large number of pro-inflammatory factors and chemokines, which aggravate the inflammatory reaction (5). In the late stage of sepsis, immune cell inactivation and endotoxin tolerance mediated by changes in immune cell phenotype, decreased antigen presentation and increased release of anti-inflammatory factors cause immunosuppression, which makes the host susceptible to secondary infection and increases mortality (6). Therefore, recognition and appropriate management of sepsis in its early stages are critical for improving outcomes (7). Sepsis sometimes exhibits a high inflammatory response pattern, followed by an immunosuppressive period during which multiple organ dysfunction may occur. Therefore, biomarkers that can reflect the immune status of sepsis patients may be a new way to predict, identify, or provide new methods for treating sepsis (8).
Autophagy is a homeostatic process by which damaged or denatured proteins, damaged organelles, or pathogens are engulfed by autophagosomes in cells and then fused with lysosomes and degraded (9). Autophagy is a key host defense mechanism against pathogens, plays a vital part in the induction and regulation of innate immune cell inflammatory responses, and influences the development of sepsis (10). Autophagy may play a protective role in sepsis through the direct clearance of pathogens, the neutralization of microbial toxins, the modulation of cytokine release, and the promotion of antigen presentation (11,12). In addition, activation of autophagy in sepsis patients can induce the formation of neutrophil extracellular traps (NETs) (13), thereby alleviating damage to host organs. Apoptosis of CD4+ T cells is an important cause for the suppression of immune function in the pathogenesis of sepsis. Increased expression of the autophagy-negative regulator Mitofusin 2 (Mfn2) suppresses immune function during sepsis by inhibiting autophagy through increasing the apoptosis of CD4+ T cells (14). Thus, autophagy plays an important role in immune regulation in patients with sepsis. However, compared with apoptosis, the autophagy-related genes (ARGs) of sepsis and their regulatory relationships with the immune regulation remain largely unknown and require further exploration. Autophagy does not always play a protective role in the development of sepsis. In acute lung injury caused by sepsis, moderate autophagy induced by oxidative stress reduces epithelial cell death, and excessive autophagy leads to increased programmed cell death (15,16). Previous studies have shown that autophagy-related genes have a significant impact on the immune cell infiltration of diseases, and can be used as potential biomarkers for diagnosis (17,18). Therefore, it is of great significance to study the potential ARGs related to the immune regulation of sepsis.
To gain insight into the regulatory function of autophagy in the immune system of patients with sepsis, this study used weighted gene co-expression network analysis machine learning to identify molecular diagnostic markers related to autophagy and determine the relationship between diagnostic markers and differential immune cells with diagnostic value in sepsis. In addition, a competing endogenous (ceRNA) network was used to clarify the underlying molecular regulatory mechanisms in sepsis. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-312/rc).
Methods
ARGs and sepsis patient data sets
We downloaded the messenger RNA (mRNA) expression profile data sets of GSE28750 and GSE95233 from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). GSE95233 data were derived from the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). The GSE28650 data set included 10 sepsis and 20 control whole blood samples. The GSE95233 data set included 102 sepsis and 22 control whole blood samples. Sepsis patients met the 1992 Consensus Statement criteria and had clinical evidence of systemic infection based on microbiology diagnoses. Moreover, 328 ARGs were obtained from the Human Autophagy Database (HADb; http://www.autophagy.lu) and the Molecular Signatures Database v. 7.1 (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
Differentially expressed analysis of the ARGs
We downloaded the expression matrix GSE28750 and GSE95233 data sets from the GEO database. After annotating the expression matrix, differentially expressed ARGs were screened out with the “limma” package in R software (The Foundation for Statistical Computing). Absolute log2 (fold change) >0.5 was set as the cutoff standard, and a P value <0.05 was considered statistically significant. A heatmap and volcano plot were used to present the results with the “heatmap” and “ggplot2” packages in R.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of ARGs and hub ARGs were conducted using the “clusterProfiler” package in R to predict their underlying molecular functions. A P value <0.05 was considered statistically significant.
Weighted Gene Coexpression Network Analysis (WGCNA)
A coexpression network for ARGs was constructed with the “WGCNA” package in R. Hierarchical clustering analysis was used to detect the outliers. To identify the critical modules related to sepsis, the soft-thresholding power was set to β=18 (scale-free R2=0.82). The modules were divided by the dynamic cutting tree algorithm, and the minModuleSize parameter in “WCGNA” was set to 10; correlations between clinical traits and sample expression were determined using the “cor” function, while the “corPvalueStudent” function was used to calculate the P value. Modules that had the highest correlation with traits and a P value <0.05 were selected. The biological function of the selected module was examined with GO and KEGG pathway analysis.
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes online database (STRING; https://string-db.org) was used to construct a PPI network. A medium confidence score of ≥0.4 was considered statistically significant. Cytoscape was used to visualize the molecular interaction networks of the STRING analysis.
Identification of hub genes
The scores of 5 ranking methods of the cytoHubba plugin of Cytoscape, including degree, maximum neighborhood component, radiality centrality, stress centrality, and closeness centrality, were computed to screen out the top 10 genes of each method. Then, by overlapping the top 10 genes of each method, the hub genes were identified. Next, the biological function of hub genes was analyzed using GO and KEGG pathway analysis.
Diagnostic value of the diagnostic biomarkers
The Wilcoxon test was conducted to verify the expression levels of hub ARGs in the 2 data sets, GSE28750 and GSE95322, with P value <0.05 being considered statistically significant. To test the predictive value of the identified biomarkers, a receiver operating characteristic (ROC) curve was generated using the mRNA expression data from GSE28750 and GSE95322 with R language packages “pROC”. The criterion for a gene to be considered a diagnostic biomarker was an area under the ROC curve (AUC) value ≥0.7.
Evaluation of immune cell subtype distribution
Immune cell subtype distribution in the blood between a sepsis and control group was evaluated by applying the CIBERSORT algorithm to LM22, a reference set with 22 immune cell subtypes. Differential immune cells were then identified with the Wilcoxon test. Least absolute shrinkage and selection operator (LASSO) regression was further performed, and the differential immune cells were identified via the overlapping results of the 2 methods. A P value <0.05 was considered to indicate statistical significance. The results were visualized using the R packages “vioplot”, “ggplot2”, and “glment”.
Correlation analysis between biomarkers and immune cells
Spearman rank correlation analysis in R software was used to determine the correlations of the identified biomarkers with the distribution of immune cells. Correlations with P values <0.05 & |cor| >0.5 were screened out and visualized using the R package “ggplot2”.
Prediction of related long noncoding RNAs (lncRNAs)
The ARGs, considered as biomarkers, were input into the miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and miRDB database to predict their targeted microRNAs (miRNAs). The target gene binding region was set as the 3' untranslated region (UTR), and a P value <0.05 was considered statistically significant. Following this, miRNet (https://www.mirnet.ca/miRNet/home.xhtml) and starBase were used to predict the upstream lncRNAs by uploading the selected miRNAs. The results obtained from the intersection were further visualized with Cytoscape.
Statistical analysis
Statistical analyses were performed using R software (version 3.6.2). Gene expression levels of our clinical samples were compared using the Student’s t-test. A P value <0.05 was considered statistically significant.
Results
Identification of differentially expressed ARGs (DEARGs) in sepsis
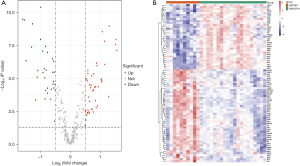
We identified 328 ARGs from the MSigDB database and the HADb database, and 317 of these were present in the GSE28750 data set. Next, we analyzed the expression of 317 ARGs of the samples in the GSE28750 data set and identified 80 DEARGs, including 47 upregulated ARGs and 33 downregulated ARGs (Table S1). Finally, the 80 DEARGs between the sepsis and control groups were visualized in a heatmap and volcano plot (Figure 1A,1B).
Function enrichment for the DEARGs
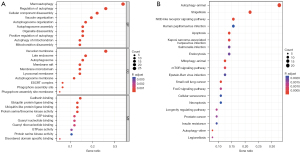
We performed GO and KEGG pathway analysis to identify the potential biological functions of the 80 DEARGs, and a total of 564 biological processes (BPs), 60 cellular components (CC), 41 molecular functions (MFs), and 55 KEGG signaling pathways were obtained. We used the R language package “ggplot2” to visualize the results, and subsequently identified the top 10 enriched GO terms and the top 30 enriched KEGG signaling pathways (Figure 2A,2B). The results showed that the most significantly enriched GO terms involved macroautophagy, autophagy regulation, and autophagosome organization. Meanwhile, the KEGG pathway enrichment analysis indicated that the DEARGs were mostly involved in autophagy, shigellosis, mitophagy, apoptosis, and NOD-like receptor signaling pathway.
Coexpression module construction
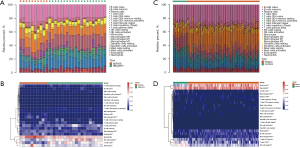
The expression matrix of the 80 DEARGs was used as input data and was assessed with the “WGCNA” package to construct a coexpression network. Clustering analyses were performed and detected no outlier (Figure 3A). The DEARGs were incorporated into the modules, and 4 modules were constructed (Figure 3B). The genes that did not belong to any module were collected in the gray module and were not used in any subsequent analyses. The other 3 modules are shown in blue, turquoise, and brown, respectively (Figure 3C). Among the 3 modules, the brown module containing 17 DEARGs had significant positive relevance for sepsis (correlation =0.82 and P<2e–8).

Identification of hub genes
PPI networks were constructed to analyze the 17 DEARG interactions in the brown module. The results revealed that the 17 DEARGs interacted with each other (Figure 4A). Then, the scores of 5 ranking methods were computed separately using the cytoHubba plugin of Cytoscape to select the hub genes. The top 10 genes of each method were extracted and identified as hub genes. By overlapping the top 10 genes of each method, 7 hub DEARGs were obtained: γ-aminobutyric acid (GABA) receptor-associated protein-like 2 (GABARAPL2), WD repeat domain, phosphoinositide-interacting 1 (WIPI1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), (WD repeat and FYVE domain containing 3 (WDFY3), DNA damage–regulated autophagy modulator 1 (DRAM1), microtubule-associated protein 1 light chain 3 β (MAP1LC3B), and Unc-51-like kinase 3 (ULK3) (Figure 4B).
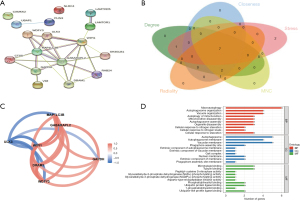
Identification and analysis of biomarkers
The R package “corrr” was used to determine the correlation of the 7 DEARGs. The results revealed that GABARAPL2, GAPDH, WDFY3, MAP1LC3B, DRAM1, and WIPI1 had positive correlations and negative correlations with ULK3 (Figure 4C). GO analysis was the performed, and 38 BPs, 13 CCs, and 25 MFs were obtained. The top 10 GO results are displayed in Figure 4D and showed that the hub DEARGs mostly involved in macroautophagy, autophagosome organization, and autophagy of mitochondrion.
Validation of biomarkers
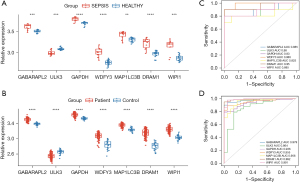
The Wilcoxon test was conducted to verify the expression levels of the 7 hub DEARGs of the samples in the GSE28750 and GSE95322 data sets. The results indicated that the samples’ mRNA expression of the 7 hub DEARGs between the sepsis and the control groups in the 2 data sets had significant differences (Figure 5A,5B). The diagnostic ability of the 7 hub DEARGs was then validated with a ROC curve using the mRNA expression data from the GSE28750 data set. This yielded AUCs of 0.885 (95% CI: 0.722–1) in GABARAPL2, 0.88 (95% CI: 0.716–1) in ULK3, 0.93 (95% CI: 0.791–1) in GAPDH, 0.985 (95% CI: 0.952–1) in WDFY3, 0.825 (95% CI: 0.606–1) in MAP1LC3B, 0.95 (95% CI: 0.849–1) in DRAM1, and 0.885 (95% CI: 0.726–1) in WIPI1. The diagnostic ability of the 7 hub DEARGs was validated in the GSE95322 data set. This yielded AUCs of 0.979 (95% CI: 0.958–0.999) in GABARAPL2, 0.864 (95% CI: 0.772–0.955) in ULK3, 0.936 (95% CI: 0.894–0.978) in GAPDH, 0.992 (95% CI: 0.922–0.988) in WDFY3, 0.956 (95% CI: 0.918–0.995) in MAP1LC3B, 0.992 (95% CI: 0.98–1) in DRAM1, and 0.991 (95% CI: 0.977–1) in WIPI1 (Figure 5C,5D). The AUC values of these ARGs were all above 0.85, indicating that the 7 hub ARGs had diagnostic values with excellent specificity and sensitivity and were determined to be diagnostic markers.
Immune infiltration analysis
The distribution of the 22 types of infiltrated immune cells in sepsis and control groups samples of the GSE28750 data set and GSE95233 data set were examined with CIBERSORT algorithm and is presented in Figure 6A-6D. The results of the Wilcoxon test conducted in GSE28750 data set are shown in a violin diagram in Figure 7A. The proportions of resting natural killer (NK) cells, CD8 T cells, naïve CD4 T cells, and activated CD4 memory T cells in the sepsis group p were significantly decreased compared with those in the control group. However, the proportions of plasma cells, monocytes, M0 macrophages, M2 macrophages, activated dendritic cells, eosinophils, and neutrophils in the sepsis group were significantly increased compared with those in the control group. The Wilcoxon test was also used to verify the composition of infiltrated immune cells of samples in the GSE95233 data set. The Wilcoxon test was also used to verify the composition of infiltrated immune cells of samples in the GSE95233 data set. As shown in Figure 7B, the proportions of naïve B cells, CD8 T cells, naïve CD4 T cells, resting NK cells, follicular helper T cells, M1 macrophages, and M2 macrophages in the sepsis group were significantly lower than those in the control group. The proportions of B cells memory, gamma-delta T cells, plasma cells, monocytes, M0 macrophages, activated dendritic cells, eosinophils, and neutrophils in the sepsis group were significantly increased compared with those in the control group.
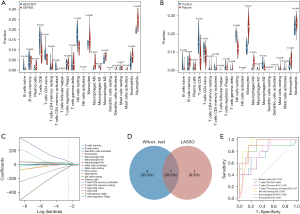
We subsequently performed LASSO regression in GSE28750 data set, and 12 significantly different types of immune cells were obtained (Figure 7C). The 2 methods extracted 7 intersecting immune cells, which were plasma cells, CD8 T cells, naïve CD4 T cells, activated CD4 memory T cells, M0 macrophages, resting NK cells, and eosinophils (Figure 7D). ROC curve analysis further verified the diagnostic value of differential immune cells. This yielded AUCs of 0.753 (95% CI: 0.54–0.965) in plasma cells, 0.865 (95% CI: 0.683–1) in T cells CD8, 0.785 (95% CI: 0.616–0.954) in T cells CD4 naive, 0.87 (95% CI: 0.74–1) in T cells CD4 memory activated, 0.935 (95% CI: 0.845–1) in NK cells resting, 0.925 (95% CI: 0.834–1) in macrophages M0, and 0.835 (95% CI: 0.681–0.989) in eosinophils. The result showed that the 7 differential immune cells had diagnostic values with AUCs >0.75 (Figure 7E).
Relationship between diagnostic biomarkers and immune cell infiltration
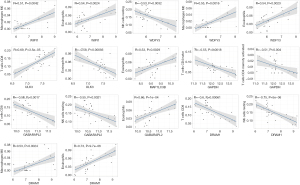
As shown in Table S2, GABARAPL2, GAPDH, WDFY3, MAP1LC3B, DRAM1, and WIPI1 were positively correlated with plasma cells, M0 macrophages, and eosinophils and negatively correlated with CD8 T cells, naïve CD4 T cells, activated CD4 memory T cells, and resting NK cells. However, ULK3 was positively correlated with CD8 T cells, naïve CD4 T cells, activated CD4 memory T cells, and resting NK cells and negatively correlated with plasma cells, M0 macrophages, and eosinophils. Correlations of biomarkers and immune cells with R>0.50 and P<0.05 were considered to be significant and were screened, with the results being presented in Figure 8.
Prediction of related lncRNAs
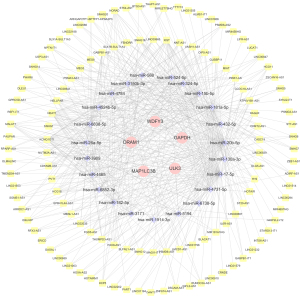
As shown in Figure 9, 72 upstream miRNAs targeting 5 hub ARGs were predicted with miRWalk and miRDB. In addition, 122 lncRNAs potentially related to the selected miRNAs were predicted.
Discussion
Immune dysfunctions, such as early immune system hyperactivation and late immunosuppression, are central pathogenic mechanisms in the development of sepsis (4). In addition, studies have shown that autophagy negatively regulates inflammatory responses and reduces inflammatory damage to various tissues and organs in sepsis (19-21).
In this study, we used the bioinformatics analysis to screen out ARGs and identified hub genes using WGCNA analysis. In all, 80 DEARGs were identified, including 47 upregulated and 33 downregulated genes, and 7 hub ARGs were identified as diagnostic biomarkers. Macroautophagy, autophagosome organization, and autophagy were the main biological functions of the hub ARGs, which were confirmed by functional enrichment analysis. MAP1LC3B is the most studied Atg8/LC3 family protein associated with autophagosome development and maturation and is used to monitor autophagic activity (22). A previous study revealed that MAP1LC3B can regulate NALP3-dependent inflammation by preserving mitochondrial integrity (11). Furthermore, in sepsis mice, overexpression of MAP1LC3 increased autophagosome clearance to alleviate lung injury and improve survival (23). GABARAPL2 also belongs to the Atg8/LC3 family, which is essential for autophagy (24). Studies indicate that GABARAPL2 plays a critical function in suppressing guanylate binding protein-dependent caspase-11–induced inflammation and septic shock (25,26). Meanwhile, WDFY3 takes part in the clearance of aggregated proteins through autophagy by interacting with the ubiquitin-binding autophagy receptors p62/SQSTM1 and NBR1 (27,28). WIPI1 contributes to autophagosome assembly and binds phosphoinositides, which are essential components of any membrane (29). One study reported that autophagosome formation can be monitored by detecting WIPI1 mRNA in a variety of cells in a wide range of cell types (30). DRAM1 is a TP53 target gene, which is involved in encoding lysosomal membrane protein. DRAM1 plays a key role in autophagy activation and apoptosis (31). Although there is no report on the role of DRAM1 in sepsis, it has been previously reported that DRAM1 activates selective autophagy in zebrafish and in human macrophage models of mycobacterial infection and plays a role in recognizing pathogens through an innate immune sensing pathway (32). Other research suggests that DRAM1 regulates autophagy via lysosomal membrane permeabilization of HIV-infected CD4 T cells, indicating the self-defense function of DRAM1 in host–pathogen interactions (33). GAPDH is a crucial cytosolic enzyme in glycolysis and the most frequently used housekeeping gene due to its stable and constitutively high expression levels in most cells (34). However, evidence indicates that increased oxidative stress might lead to increased GAPDH expression in sepsis whole blood samples (35). Meanwhile, one study reported that phosphorylation of GAPDH by activated AMPK caused GAPDH to redistribute into the nucleus and increased Sirt1 activation and autophagy upon glucose starvation (36). Another study found that GAPDH preinjection had an anti-inflammatory effect and prolonged survival in a liposaccharide-induced ALI mouse model (37). In a sepsis mouse model, GAPDH was found to regulate the immunomodulation on macrophage functions and polarization (38). However, no literature on ULK3 and its functions in sepsis yet exists. ULK3 is considered to be part of noncanonical upstream regulatory complex FP200/ATG13/ULK, which is required for LC3C autophagic programming in clear cell renal cell carcinoma (39). Studies have indicated that increased ULK3 can induce autophagy in human diploid fibroblasts and squamous cell carcinoma (40,41). However, our study found that the expression of ULK3 decreased while the other autophagy markers increased and that ULK3 was negatively correlated with the other hub ARGs. Clarifying the role of ULK3 in sepsis requires further study.
ceRNA is an element that can compete for binding mRNA. miRNAs and lncRNAs are 2 kinds of ceRNAs. miRNAs are a class of RNA molecules that can bind to target mRNAs, while some lncRNAs can competitively bind to miRNAs, reducing the binding effect of miRNAs. Binding of miRNAs to target mRNAs can lead to mRNA degradation or translation to regulate gene expression (42). An increasing amount of research suggests that ceRNAs occupy an essential role in the development and the autophagy process of sepsis. Wang et al. found that miR-20a can activate autophagy and promote the progression of kidney injury in septic rats (43). Gui et al. found that increased circulating lnc-ANRIL-miR-125a axis level could indicate a worse prognosis of sepsis (44). We constructed an mRNA-miRNA-lncRNA ceRNA network to predict the interactions of a potential ceRNA network targeting hub ARGs that may influence the autophagy of sepsis. The network may provide novel biomarkers and promising therapeutic targets, but further research is needed to confirm its value.
Multiple immune cells play an essential role in sepsis: neutrophils migrate to the site of infection to exert phagocytosis and bactericidal action (45); dendritic cells are the most functionally proficient antigen-presenting cells (APCs) which can efficiently take up, process, and present antigens (46); T cells participate in adaptive immune response (47,48); and NK cells perform nonspecific direct killing of the pathogen (49). This study used 2 machine learning methods to assess the type of immune cell infiltration in the sepsis and control samples, and multiple differential immune cell subtypes were found. The result showed that the proportions of CD8 T cells, naïve CD4 T cells, activated CD4 memory T cells, and resting NK cells in the sepsis group were significantly lower than those in the control group, while the proportions of plasma cells, M0 macrophages and eosinophils in the sepsis group were significantly higher than those in the control group. We also conducted a Wilcoxon test to explore the distribution of immune cells in the GSE95233 data set. The results showed that the proportion of resting NK cells in the sepsis group was significantly decreased compared with that in the control group. The proportions of M0 Macrophages and eosinophils in the sepsis group were significantly increased compared with those in the control group, consistent with previous analysis. However, the other differential immune cells were different from the previous analysis. This discrepancy may be due to the different immune statuses in the development of sepsis in the different data sets. In the late stage of sepsis, a mass death of immune cells, especially macrophages, may cause immunosuppression and deterioration of the infectious states (50). In this study, correlation analysis indicated that all hub ARGs except ULK3 were positively correlated to the differential immune cells, including plasma cells, M0 macrophages, and eosinophils, indicating that these ARGs may exert protective effects on some immune cells through autophagy activation. Eosinophilia may indicate an immune imbalance due to type 2 inflammation that activates eosinophils. Moreover, eosinophil consumption may be responsible for eosinophilia, which can contribute to dysregulated host responses in infection and acute respiratory distress syndrome. Persistent peripheral eosinopenia was found to be independently associated with poor outcomes in sepsis (51). Therefore, the high expression of ARGs may cause some immune cells to activate in the early stage of sepsis, leading to the immune-activated microenvironment. Regulating the expression of ARGs may become a treatment for organ function damage caused by cytokine storm in early stage of sepsis, which may help guide immune modulators to achieve immune homeostasis. A previous study indicated that autophagy could prevent monocytes from undergoing apoptosis and promote their differentiation into macrophages (52). Induction of autophagy can alleviate the damage of excessive inflammatory injury by inducing the transition of macrophages from the M1 phenotype to the M2 phenotype (53). Moreover, some immune cells, including CD8 T cells, naïve CD4 T cells, activated CD4 memory T cells, and resting NK cells, have been reported to be negatively correlated with hub ARGs. In contrast to our findings, one study found that autophagy deficiency in T cells was associated with increased apoptosis of CD4+ and CD8+ (54). The effect of autophagy on the immune regulation of sepsis still needs further study. The above studies and our current findings suggest that autophagy plays an essential role in sepsis immune regulation and should be the focus of future research.
Our study still has some limitations. First, to ensure the homogeneity of the samples in the data set, we used a single data set for analysis, while the GSE28750 data set has a small number of cases. Second, we only used database data without clinical sample experimental validation, which is a limitation. Third, for the analysis of the immune microenvironment in sepsis, we do not have data to support the analysis of the patients with different immune statuses. Therefore, more studies should be performed to validate our conclusions.
Conclusions
We identified 80 potential DEARGs of sepsis through the bioinformatics analysis. GABARAPL2, GAPDH, WDFY3, MAP1LC3B, DRAM1, and WIPI1 were identified as biomarkers and may influence the development of sepsis by regulating autophagy. We also constructed a ceRNA network targeting biomarkers and predicted 23 miRNAs and 122 lncRNAs regulating 5 biomarkers. Plasma cells, CD8 T cells, CD4 naïve T cells, CD4 memory activated T cells, resting NK cells, M0 Macrophages, and eosinophils may have diagnostic value for sepsis and correlate with autophagy. Regulating the expression of ARGs may help guide immune modulators to achieve immune homeostasis.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81871540), the Clinical Research Plan of SHDC (Nos. SHDC2020CR6030-003, SHDC2020CR1028B, and SHDC12020122), the Three-Year Action Plan for the Construction of the Shanghai Public Health System (No. GWV-3.1), the Construction Plan of Important and Weak Disciplines of the Shanghai Health Commission (No. 2016ZB0204-01), and the Shanghai Science and Technology Innovation Action Plan (No. 18ZR1429500).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-312/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-312/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-312/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- Fleischmann-Struzek C, Mellhammar L, Rose N, et al. Incidence and mortality of hospital- and ICU-treated sepsis: results from an updated and expanded systematic review and meta-analysis. Intensive Care Med 2020;46:1552-62. [Crossref] [PubMed]
- Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet 2020;395:200-11. [Crossref] [PubMed]
- Delano MJ, Ward PA. The immune system's role in sepsis progression, resolution, and long-term outcome. Immunol Rev 2016;274:330-53. [Crossref] [PubMed]
- Lauvau G, Loke P, Hohl TM. Monocyte-mediated defense against bacteria, fungi, and parasites. Semin Immunol 2015;27:397-409. [Crossref] [PubMed]
- Lee CR, Zeldin DC. Resolvin Infectious Inflammation by Targeting the Host Response. N Engl J Med 2015;373:2183-5. [Crossref] [PubMed]
- Evans L, Rhodes A, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med 2021;47:1181-247. [Crossref] [PubMed]
- Liu YC, Shou ST, Chai YF. Immune checkpoints in sepsis: New hopes and challenges. Int Rev Immunol 2022;41:207-16. [Crossref] [PubMed]
- Vargas JNS, Hamasaki M, Kawabata T, et al. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol 2023;24:167-85. [Crossref] [PubMed]
- Wang R, Xu Y, Fang Y, et al. Pathogenetic mechanisms of septic cardiomyopathy. J Cell Physiol 2022;237:49-58. [Crossref] [PubMed]
- Nakahira K, Haspel JA, Rathinam VA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011;12:222-30. [Crossref] [PubMed]
- Ryter SW, Mizumura K, Choi AM. The impact of autophagy on cell death modalities. Int J Cell Biol 2014;2014:502676. [Crossref] [PubMed]
- Huang Z, Zhang H, Fu X, et al. Autophagy-driven neutrophil extracellular traps: The dawn of sepsis. Pathol Res Pract 2022;234:153896. [Crossref] [PubMed]
- Ying L, Zhao GJ, Wu Y, et al. Mitofusin 2 Promotes Apoptosis of CD4(+) T Cells by Inhibiting Autophagy in Sepsis. Mediators Inflamm 2017;2017:4926205. [Crossref] [PubMed]
- Tanaka A, Jin Y, Lee SJ, et al. Hyperoxia-induced LC3B interacts with the Fas apoptotic pathway in epithelial cell death. Am J Respir Cell Mol Biol 2012;46:507-14. [Crossref] [PubMed]
- Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014;157:65-75. [Crossref] [PubMed]
- He S, Deng Z, Li Z, et al. Signatures of 4 autophagy-related genes as diagnostic markers of MDD and their correlation with immune infiltration. J Affect Disord 2021;295:11-20. [Crossref] [PubMed]
- Tan G, Wu A, Li Z, et al. Bioinformatics analysis based on immune-autophagy-related lncRNAs combined with immune infiltration in bladder cancer. Transl Androl Urol 2021;10:3440-55. [Crossref] [PubMed]
- Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008;456:264-8. [Crossref] [PubMed]
- Waltz P, Carchman EH, Young AC, et al. Lipopolysaccaride induces autophagic signaling in macrophages via a TLR4, heme oxygenase-1 dependent pathway. Autophagy 2011;7:315-20. [Crossref] [PubMed]
- Sun Y, Yao X, Zhang QJ, et al. Beclin-1-Dependent Autophagy Protects the Heart During Sepsis. Circulation 2018;138:2247-62. [Crossref] [PubMed]
- Schaaf MB, Keulers TG, Vooijs MA, et al. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J 2016;30:3961-78. [Crossref] [PubMed]
- Lo S, Yuan SS, Hsu C, et al. Lc3 over-expression improves survival and attenuates lung injury through increasing autophagosomal clearance in septic mice. Ann Surg 2013;257:352-63. [Crossref] [PubMed]
- Bento CF, Renna M, Ghislat G, et al. Mammalian Autophagy: How Does It Work? Annu Rev Biochem 2016;85:685-713. [Crossref] [PubMed]
- Eren E, Planès R, Bagayoko S, et al. Irgm2 and Gate-16 cooperatively dampen Gram-negative bacteria-induced caspase-11 response. EMBO Rep 2020;21:e50829. [Crossref] [PubMed]
- Sakaguchi N, Sasai M, Bando H, et al. Role of Gate-16 and Gabarap in Prevention of Caspase-11-Dependent Excess Inflammation and Lethal Endotoxic Shock. Front Immunol 2020;11:561948. [Crossref] [PubMed]
- Clausen TH, Lamark T, Isakson P, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010;6:330-44. [Crossref] [PubMed]
- Filimonenko M, Isakson P, Finley KD, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 2010;38:265-79. [Crossref] [PubMed]
- Müller AJ, Proikas-Cezanne T. Function of human WIPI proteins in autophagosomal rejuvenation of endomembranes? FEBS Lett 2015;589:1546-51. [Crossref] [PubMed]
- Tsuyuki S, Takabayashi M, Kawazu M, et al. Detection of WIPI1 mRNA as an indicator of autophagosome formation. Autophagy 2014;10:497-513. [Crossref] [PubMed]
- Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy 2007;3:72-4. [Crossref] [PubMed]
- van der Vaart M, Korbee CJ, Lamers GE, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense Cell Host Microbe 2014;15:753-67. [corrected]. [Crossref] [PubMed]
- Laforge M, Limou S, Harper F, et al. DRAM triggers lysosomal membrane permeabilization and cell death in CD4(+) T cells infected with HIV. PLoS Pathog 2013;9:e1003328. [Crossref] [PubMed]
- Barber RD, Harmer DW, Coleman RA, et al. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 2005;21:389-95. [Crossref] [PubMed]
- Cummings M, Sarveswaran J, Homer-Vanniasinkam S, et al. Glyceraldehyde-3-phosphate dehydrogenase is an inappropriate housekeeping gene for normalising gene expression in sepsis. Inflammation 2014;37:1889-94. [Crossref] [PubMed]
- Chang C, Su H, Zhang D, et al. AMPK-Dependent Phosphorylation of GAPDH Triggers Sirt1 Activation and Is Necessary for Autophagy upon Glucose Starvation. Mol Cell 2015;60:930-40. [Crossref] [PubMed]
- Takaoka Y, Goto S, Nakano T, et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Sci Rep 2014;4:5204. [Crossref] [PubMed]
- Nakano T, Goto S, Takaoka Y, et al. A novel moonlight function of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for immunomodulation. Biofactors 2018;44:597-608. [Crossref] [PubMed]
- Bischoff ME, Zang Y, Chu J, et al. Selective MAP1LC3C (LC3C) autophagy requires noncanonical regulators and the C-terminal peptide. J Cell Biol 2021;220:e202004182. [Crossref] [PubMed]
- Young AR, Narita M, Ferreira M, et al. Autophagy mediates the mitotic senescence transition. Genes Dev 2009;23:798-803. [Crossref] [PubMed]
- Goruppi S, Procopio MG, Jo S, et al. The ULK3 Kinase Is Critical for Convergent Control of Cancer-Associated Fibroblast Activation by CSL and GLI. Cell Rep 2017;20:2468-79. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Wang JX, Tao YL, Wang Z, et al. MiR-20a Promotes kidney injury in sepsis rats through autophagy. J Biol Regul Homeost Agents 2020;34:1277-83. [PubMed]
- Gui F, Peng H, Liu Y. Elevated circulating lnc-ANRIL/miR-125a axis level predicts higher risk, more severe disease condition, and worse prognosis of sepsis. J Clin Lab Anal 2019;33:e22917. [Crossref] [PubMed]
- Kovach MA, Standiford TJ. The function of neutrophils in sepsis. Curr Opin Infect Dis 2012;25:321-7. [Crossref] [PubMed]
- Pastille E, Didovic S, Brauckmann D, et al. Modulation of dendritic cell differentiation in the bone marrow mediates sustained immunosuppression after polymicrobial sepsis. J Immunol 2011;186:977-86. [Crossref] [PubMed]
- Rimmelé T, Payen D, Cantaluppi V, et al. Immune cell phenotype and function in sepsis. Shock 2016;45:282-91. [Crossref] [PubMed]
- Luan YY, Dong N, Xie M, et al. The significance and regulatory mechanisms of innate immune cells in the development of sepsis. J Interferon Cytokine Res 2014;34:2-15. [Crossref] [PubMed]
- Giamarellos-Bourboulis EJ. Natural killer cells in sepsis: detrimental role for final outcome. Crit Care Med 2014;42:1579-80. [Crossref] [PubMed]
- Qiu P, Liu Y, Zhang J. Review: the Role and Mechanisms of Macrophage Autophagy in Sepsis. Inflammation 2019;42:6-19. [Crossref] [PubMed]
- Al Duhailib Z, Farooqi M, Piticaru J, et al. The role of eosinophils in sepsis and acute respiratory distress syndrome: a scoping review. Can J Anaesth 2021;68:715-26. [Crossref] [PubMed]
- Zhang Y, Morgan MJ, Chen K, et al. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood 2012;119:2895-905. [Crossref] [PubMed]
- Ip WKE, Hoshi N, Shouval DS, et al. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 2017;356:513-9. [Crossref] [PubMed]
- Lin CW, Lo S, Hsu C, et al. T-cell autophagy deficiency increases mortality and suppresses immune responses after sepsis. PLoS One 2014;9:e102066. [Crossref] [PubMed]
(English Language Editor: J. Gray)