
Cyclin O promotes lung cancer progression and cetuximab resistance via cell cycle regulation and CDK13 interaction
Highlight box
Key findings
• CCNO interacts with CDK13 to increase EGFR signaling and promote cetuximab tolerance.
What is known, and what is new?
• CCNO is a proto-oncogene that plays a role in breast, lung, and stomach cancers. CCNO knockdown can induce tumor cell apoptosis in gastric and lung cancer.
• CCNO activates the EGFR signaling pathway. CCNO interacts with CDK13.
What is the implication, and what should change now?
• Target drug tolerance in patients with high CCNO expression may be related to the interaction between CCNO and CDK13.
Introduction
Lung adenocarcinoma (LUAD) is the most prevalent form of lung cancer and one of the leading causes of cancer-related mortality globally (1). Patients with LUAD continue to have high recurrence rates and low survival despite substantial efforts to improve early detection and develop novel treatment options. Therefore, the search for additional LUAD driver genes should be prioritized to provide new diagnostic and targeted therapy for LUAD. Several proteins, cirRNA, and mRNA have been published to indicate their association with the prognosis of LUAD, but there is little further exploration of their molecular mechanisms. At the same time, there is only a prompt and warning effect for LUAD patients, and there is no guiding significance for clinical treatment.
The capability for sustaining proliferative signaling is one of the hallmarks of cancer, and its primary molecular mechanism is cell cycle disruption (2). Cell cycle disorders are also central to genomic instability, which can lead to further oncogenesis of unstable cells and thus participate in tumor development. The cell cycle is a complex regulatory process involving multiple genes, including three classes of sophisticated regulators, namely cyclin-dependent kinases (CDKs), cyclins, and cyclin-dependent kinase inhibitors (CKIs) , of which CDKs are at the center of cell cycle regulation, while cyclins activate CDK and thus regulate the cell cycle through periodic changes in expression. The eukaryotic cell cycle is regulated by the temporal activation of different CDK/cyclin complexes. Through these major transitions is mediated by sequential activation and inactivation of CDKs, which is frequently accompanied by phosphorylation of serine and threonine often occurs (3). Meanwhile, the cyclin activation and the repression of CKIs proteins can be regulated by EGFR signal pathway (4). Cyclin O (CCNO) is a novel cyclin family protein containing a cyclin-like domain, consisting of three exons located on chromosome 5q11 encoding a 1,053 nt mRNA and a 350-aminoacid protein (5). According to reports, CCNO is produced during the S phase and co-localizes with the replication foci of proliferating cell nuclear antigen protein before being destroyed during the S/G2 transition (6). Additionally, CCNO is linked to DNA damage-induced apoptosis in mouse lymphoid cells. CCNO has been demonstrated to bind to CDK2 to create the CCNO-CDK1/CDK2 complex, which regulates the cell cycle and apoptotic signals (7). CDK2 is the predominant activating complex form of CCNO, but CCNO can bind to CDK1 to form an activating complex in the absence of CDK2. Ma et al. revealed that CCNO is present in oocytes and may play a role in oocyte maturation and development (8). The role of CCNO in malignancies has not been well investigated. CCNO is abundantly expressed in stomach and cervical cancer and, according to previous reports, its downregulation induces apoptosis in tumor cells (9,10). However, neither the expression nor the function of CCNO in lung cancer has been previously described. In previous studies, the study of CCNO was limited to the differential expression of its protein level in cancer, such as gastric cancer and cervical cancer. Further functional research has not been involved, let alone exploration of its molecular mechanisms. Studying the relationship between CCNO and the progression of LUAD can preferably individualized targeted treatment, and researching more precise drugs to improve the targeted drug treatment plan for LUAD patients with CCNO participation.
CDKs are a set of serine-threonine protein kinases that regulate several physiological processes, including the cell cycle. They are split into two major categories: cell cycle-associated CDKs and transcription-associated CDKs (11). CDK12/13, as transcriptional regulatory CDKs, are highly expressed in a variety of tissues and regulate the expression of several genes. CDK13 has been demonstrated to play a significant role in the development of pancreatic, ovarian, and prostate cancers, where it is aberrantly expressed in several tumor types (12-14). It has been revealed that CDK12 regulates downstream signaling by phosphorylating EGFR (15), an essential driver gene and therapeutic target in lung cancer. Several small-molecule inhibitors of CDK12/13 have been discovered; however, their applicability in tumor therapy is restricted. CDK12/13 are also suitable targets for tumor therapy (16), but there is insufficient research on the CDK13 mode of action in lung cancer.
In this study, we aimed to examine whether CCNO is substantially expressed in LUAD and supports the malignant progression of the tumor phenotype. We hypothesized that CCNO might bind to CDK13 and increase EGFR downstream signaling. In addition, we explored whether CDK13 inhibitors enhanced the susceptibility of LUAD cells to cetuximab. This study was conducted to identify novel biomarkers and treatment targets for LUAD. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-437/rc).
Methods
Human LUAD samples and cell lines
Seven samples of paired fresh LUAD and adjacent normal tissues (>10 cm from the tumor tissue) were recruited from patients undergoing surgical resection in Department of Thoracic Surgery, Qilu Hospital of Shandong University between 2004 and 2010. The excised tissue samples were immediately placed into liquid nitrogen until needed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for this study was granted by the Medical Ethics Committee of Qilu Hospital of Shandong University (No. KYLL-2019(KS)-046). Informed consent was obtained from each patient or their legal guardians. Pathological examination confirmed all diagnoses. Detailed clinicopathological and observational data were available for 94 paired LUAD samples. The admission criteria for LUAD patients were mentioned in the Table 1, and we continuously telephone follow-up to track the survival time of the patients for statistical analysis. We acquired HBE, H1299, A549, PC9, H1975, and HCC827 cell lines from the Shanghai Cell Bank of the Institute of Cell Research of the Chinese Academy of Sciences, cultured in a regular 1640 medium. All cell lines underwent STR profiling and were checked every 3 months for mycoplasma contamination. Cells were transiently transfected with siRNA or plasmids using a jetPRIME transfection reagent (Polyplus, USA) according to the manufacturer’s protocol. After 48–72 h of transfection, the cells were collected and lysed to measure the transfection efficiency. CCNO overexpression and knockdown lentiviruses were purchased from Guangzhou Genecopia Company. H1299 stable cell lines overexpressing CCNO were transfected with lentiviruses (MOI: 20) and selected with 1 mg/mL puromycin for about 1 week. H1299 and A549 stable cell lines lacking CCNO were transfected with lentiviruses (MOI: 20) and selected with 0.5 and 1 mg/mL puromycin, respectively, for approximately 1 week. The siRNA sequences are shown in the Supplementary file (Appendix 1).
Table 1
| Characteristics | CCNO | P value | |
|---|---|---|---|
| Low | High | ||
| Age (years) | 0.794 | ||
| <60 | 22 | 27 | |
| ≥60 | 19 | 26 | |
| Gender | 0.639 | ||
| Male | 22 | 31 | |
| Female | 19 | 22 | |
| T classification | |||
| T1 | 16 | 10 | |
| T2 | 18 | 25 | 0.016* |
| T3 | 7 | 18 | |
| N classification | |||
| N0 | 22 | 24 | |
| N1 | 11 | 8 | 0.079 |
| N2 | 8 | 21 | |
| N3 | |||
| TNM stage | |||
| I | 16 | 13 | |
| II | 16 | 14 | 0.058 |
| III | 9 | 26 | |
| Pathologic differentiation | |||
| I | 12 | 5 | |
| II | 15 | 23 | 0.044* |
| III | 14 | 25 | |
The median age at diagnosis in patients with LUAD was 60 years. Samples were divided into two groups based on median age. *, statistical significance. CCNO, Cyclin O; LUAD, lung adenocarcinoma
Animal experiments
All animal experiments were approved by the Medical Ethics Committee of Shandong University (No. KYLL-2021(KS)-1053), in compliance with institutional guidelines for the care and use of animals. BALB/c nude mice (bought from GemPharmatech Co., Ltd., male, 6–8 weeks old) were used for the animal experiments because of LUAD cell lines injection derived from human. Their average weight was 18 g. The animals were randomly assigned, and during the experiments and evaluation of the results, experimental masking was performed to blind the researchers to the group assignments. A protocol was prepared before the study without registration.
For the subcutaneous xenograft experiments, either the H1299 cell line (3×106 cells/mouse), stable knockdown CCNO H1299 cell line, or control H1299 cells were subcutaneously injected into the nude mice.
Tumor size was measured every 5 days using a Vernier calibrator, and tumor volume was calculated using the formula: V=1/2 × d2 × D. After 4 weeks, the mice were sacrificed, and the tumors were removed for evaluation.
Western blotting
Protein from the cells or tissues was lysed using RIPA lysis buffer with a 1% protease and phosphatase inhibitor cocktail (Beyotime Biotechnology). Then, the extracted protein was resolved by 6–10% SDS-PAGE gel and transferred onto PVDF membranes.
Antibodies
The following antibodies were used: antibody against CCNO (Abcam Cat#ab47682), antibody against HER-2 (Abcam Cat#ab214275), antibody against EGFR (Abcam Cat#52894), antibody against beta Actin (Invitrogen Cat#MA1-140), antibody against CDK13 (Santa Cruz Cat#sc-81837), antibody against P-EGFR (Cell Signaling Cat#2236), antibody against p-HER2 (Cell Signaling Cat#2243), antibody against CDK12 (Cell Signaling Cat#11973), antibody against GAPDH (Cell Signaling Cat#5174), antibody against p-p70s6k (Cell Signaling Cat#97596), antibody against p70s6k (Cell Signaling Cat#9202), antibody against pAKT (Cell Signaling Cat#4060), antibody against AKT (Cell Signaling Cat#4691), and antibody against Anti-Flag Tag (Cell Signaling Cat#147935).
Co-immunoprecipitation was also used against IgG (ThermoFisher) and β-actin. Goat secondary antibodies against rabbit or mouse primary antibodies (Huabio) were used at a dilution of 1:5,000.
EdU staining
The transduced cells were inoculated at 5×104 cells/well into 96-well plates. The EdU staining was carried out with an EdU staining kit (RiboBio, Guangzhou, China) following the manual instruction with 2 h of EdU incubation. EdU imaging was conducted with fluorescence microscopy (Olympus, Tokyo, Japan).
Cell Counting Kit-8 (CCK8) assay
Transduced H1299 and A549 cells were seeded in 96-well microplates at 2,000 per well. Then, a 10% CCK8 solution was added to each well, and the cells were incubated in the dark for 2 h. The cells were cultured for 0, 24, 48, 72, and 96 h, and the absorbance of each well at 450 nm was recorded using a microplate reader. These experiments were replicated three times.
Wound healing, migration, and invasion assays
For wound healing, we seeded medium numbers of cells in six-well plates. The principle of overnight inoculation is 100% confluence. The cell layer at the bottom of the wells was scratched with a sterile 200 µL pipette tip to form a linear gap. Images were obtained by photographing the flat plate under an inverted fluorescent microscope at 24 and 36 hours, respectively. The rate of closure of open wounds was calculated. The scratch healing rate = (healing width at 24 or 36 hours−healing width at 0 hours)/healing width at 0 hours. Migration studies were performed using a 24-well plate across the well chamber (BD Biosciences, #353092). LUAD cells treated in different ways were seeded in the upper chamber and suspended in 100 µL of serum-free medium, and the lower chamber was filled with 700 µL of 1640 containing 20% fetal bovine serum (FBS). After 48 hours of incubation at 37 ℃, the cells were removed from the upper surface of the membrane with a cotton swab. Migrating cells in the lower half of the filter were fixed with 4% paraformaldehyde and were stained with 0.1% crystal violet for 60 min at room temperature. The number of cells migrating to the lower surface was counted in three randomly selected high-magnification fields for each sample. For the invasion studies, Matrigel (BD, Biocoat, No. 358248) was smeared on the surface of the upper chamber. The subsequent steps were the same as for the migration studies.
Immunohistochemistry (IHC)
The LUAD tissues and tumors of nude BALB/c nude mice were fixed with over 4% paraformaldehyde at room temperature for 60 minutes. They were incubated at 62 ℃ for 2 hours, dewaxed and rehydrated, and then placed in citrate buffer at 97 ℃ (pH 6.0) Extract the antigen for 20 minutes. Block endogenous peroxidase activity with a 3% hydrogen peroxide solution at room temperature for 10 minutes. Incubate plastic sheets and 5% normal goat serum in PBST at room temperature for 1 hour to avoid non-specific antibody binding. Then incubate glass sheets with the first anti-CDK13 and CCNO antibodies at 4 ℃ overnight for incubation. After washing with TBST three times, incubate each loading volume with a second antibody labeled with appropriate kinds of HRP. Then generate a signal using DAB solution before retesting with hydroxylamine. Analyze the intensity of IHC staining with using the application Image Pro Plus (IPP).
RNA assay
Total cellular RNA was extracted using an RNA extraction kit (RNAfast200, Fastagen Biotech Co., Shanghai, China), and reverse transcription was conducted with an AG reverse transcription kit. RT-qPCR reactions were run on the Bio-Rad IQ 5 RT-PCR detection system using SYBR Green Supermix (AG). GAPDH and β-actin were used as the endogenous controls. The primers were ordered from Biosune (Shanghai, China). All sequences are shown in the Supplementary file (Appendix 1).
Co-IP and immunoblot analysis
The entire cell contents were lysed in IP buffer [1.0% Nonidet P40 (Solarbio, N8030), 50 mM Tris HCl (Solarbio, Z9912), pH 7.4, 50 mM EDTA (Solarbio, E1170), and 150 mM NaCl] and a protease inhibitor mixture (Roche Diagnostics; GmbH, 11836170001) and solubilized. After centrifugation at 12,000 g for 10 min, the supernatant was collected and then incubated with Protein G Plus-Agarose Immunoprecipitation Reagent (Santa Cruz Biotechnology; sc-2003) and an IP antibody. After incubation overnight, the microbeads were washed four times with the immunoprecipitation buffer. The immunoprecipitates were eluted by boiling with 1% SDS sample buffer at 100 ℃ for 5 min. Equal amounts of extracts were separated by SDS-PAGE and then transferred onto nitrocellulose membranes and blotted with specific antibodies.
Colony formation assay
The cells were incubated in 6-well culture plates (1,000 cells/well) until cell colonies were visible. Then, the cells were fixed in 4% paraformaldehyde for 20 min and stained with 0.5% crystal violet for 30 min. Colonies larger than 50 cells were counted. Finally, the plates were washed with PBS and photographed with a digital camera. These experiments were replicated three times.
Statistical analysis
Each experiment involves three biological replicates instead of technical replicates. We carried out the statistical evaluations using GraphPad Prism 8.3.0 software (GraphPad Software Inc., San Diego, CA, USA). The data are represented as means ± standard deviations. Statistical differences were analyzed using the t-test, one-way analysis of variance (ANOVA), Kaplan-Meier survival analysis, or Pearson’s correlation. A P value <0.05 was defined as a statistically significant difference.
Results
CCNO expression was highly expressed in LUAD tissues and predicted the overall survival of LUAD patients
First, we screened potential biomarkers by transcriptome microarrays (GSE 140797) using seven pairs of frozen LUAD and adjacent normal tissues. Based on the microarray results, we found that CCNO was significantly more highly expressed in LUAD tissues than adjacent normal tissues (Figure 1A). To investigate the potential role of CCNO in the pathogenesis of human LUAD, we first analyzed the Gene Expression Omnibus (GEO) LUAD database (GSE19804, GSE32665, and GSE40791) and The Cancer Genome Atlas (TCGA) LUAD database (Figure 1B,1C). Similar to our microarray analysis results, we found that CCNO mRNA expression was upregulated in LUAD compared to adjacent normal tissues. We performed IHC staining to analyze the CCNO protein expression in 94 samples from patients with LUAD. IHC scoring showed that CCNO expression was significantly elevated in LUAD tissues compared to adjacent normal tissues (Figure 1D). Moreover, the expression level of CCNO was closely associated with the prognosis of patients. High CCNO expression predicted poorer survival (Figure 1E). Similarly, CCNO protein expression was higher in six LUAD samples than in their adjacent normal lung tissues (Figure 1F).
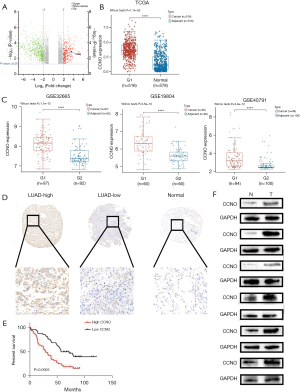
We then analyzed the relationship between clinicopathological parameters and CCNO expression. The results showed that 41 (43.6%) and 53 (56.4%) patients had lower and higher CCNO expression, respectively (Table 1). A high expression of CCNO was significantly associated with tumor stage (P=0.016, Table 1), number of lymph nodes (P=0.079, Table 1), and pathological stage (P=0.044, Table 1). However, there was no association with age, sex, or metastatic stage. In addition, Kaplan-Meier survival analysis showed that patients with higher CCNO expression had a shorter OS than those with lower expression [Supplementary file (Appendix 1), n=1,927, P<0.05]. The median survival time for patients with low CCNO expression was 107 months, while LUAD patients with high CCNO expression survived only 89 months. Taken together, our data imply an oncogenic role for CCNO in LUAD.
CCNO promoted the proliferation, migration, and invasion of LUAD cells in vitro
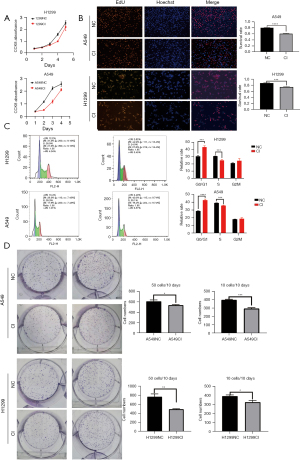
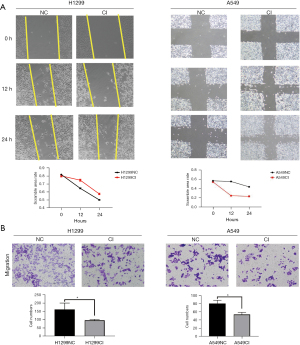
Among the seven cell lines, we found that A549 cells had a higher CCNO expression, while H1299 cells had a lower CCNO expression. Therefore, we selected H1299 and A549 cells to explore the effect of CCNO on LUAD cells [see Supplementary file (Appendix 1)]. We established CCNO-knockdown H1299 cells, A549 cells, and CCNO-overexpressing H1299 cells. Via several experiments, we found that CCNO knockdown altered the proliferation rate and the migration and invasion ability of H1299 and A549 cells. Knockdown of CCNO significantly inhibited the rate of cell proliferation and decreased cell activity. Meanwhile, CCK8 and EdU assays revealed that the survival ratio of CCNO knockdown H1299 and A549 cells was reduced compared to normal tumor cells (Figure 2A,2B). When the cell cycle was also examined, the tumor cells with knockdown CCNO were significantly blocked in the G0/G1 phase (Figure 2C), which can be the potential causes of decelerated cell proliferation. For tumor cells with knockdown CCNO, the lower CCNO cells have smaller clone number and colony size (Figure 2D). Regarding cell migration and invasion ability, tumor cells with knockdown of CCNO were weaker than normal tumor cells in terms of scratch healing area and number of migrating cells in the Transwell chambers (Figure 3A,3B).
CCNO promoted the proliferation, migration, and invasion of LUAD cells in vivo
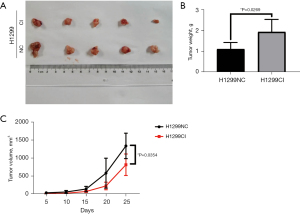
In addition, we performed in vivo experiments to assess the oncogenic effects of CCNO. We first established the knockdown of CCNO in A549 and H1299 CCNOsi (CI) cell lines. The H1299 cells were implanted subcutaneously into nude mice to establish a transplanted tumor mouse model (Figure 4A). A total of 10 male nude mice weighing about 18.0 g were selected and randomly divided equally into two groups for injection of LUAD cells with different levels of CCNO expression. The tumors which were sourced from H1299-shCCNO cells showed significantly less tumor growth and volume (Figure 4B,4C). The slowed growth volume and reduced tumor weight demonstrated the oncogenic effect of CCNO in vivo.
At the beginning of our screening, CCNO, as an oncogene, showed its oncological effect in LUAD. CCNO showed its relationship with the survival and prognosis of LUAD patients in the database and clinical sample statistics. It is our preliminary exploration that CCNO can be used as a biomarker of LUAD.
CDK13 was shown to be a downstream target gene for the oncological role of CCNO in LUAD
After demonstrating the oncological effects of CCNO, we performed high-throughput sequencing at the mRNA level in H1299 cells with knockdown of CCNO and performed biological statistics and GO analysis of the altered mRNA in the cells to obtain a variety of biological functions associated with them (Figure 5A). We found that H1299 cells with CCNO knockdown had significant alterations in their cell cycle progression and functions related to checkpoints. Meanwhile, we performed liquid-phase mass spectrometry analysis of the constructed H1299 cells overexpressing CCNO (Flag-CCNO H1299) and obtained a series of proteins that could bind with CCNO (Figure 5B). The GEO database analysis also found a weak positive correlation between CCNO expression and CDK13 (Figure 5C, r=0.122, P<0.05). Since the role of CDK13 is unknown in LUAD, we firstly found that the Pearson regression statistics (Figure 5D, r=0.3922, P<0.0001) showed CDK13 expression had a positive correlation with CCNO, leading to the same conclusion as above for mRNA. We further performed IHC experiments to stain CDK13 in 94 patients, and we found that CDK13 expression was higher in tumor tissues than in normal tissues (Figure 5E). After the previous statistical analysis of the correlations, we performed immunoprecipitation and silver staining experiments on the potentially associated proteins and identified CDK13 as a downstream binding protein of CCNO (Figure 5F,5G). Finally, intracellular immunofluorescence staining was performed, and co-stained CCNO and CDK13 were found to be co-expressed and bound in the nucleus (Figure 5H).
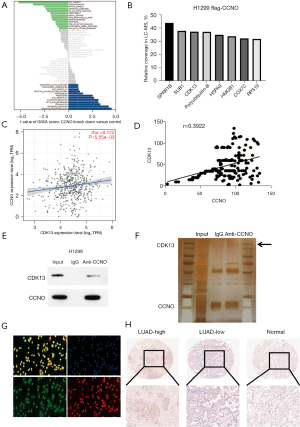
CCNO activated the P-EGFR signaling pathways via CDK13 in vitro
CDK12 and CDK13 are known to bind to each other and activate the downstream phosphorylation pathway, which leads to EGFR pathway activation and promotes the biological effects of tumor cell proliferation and survival. CDK13 is a downstream target gene of CCNO; they bind to each other and phosphorylate the EGFR pathway to exert oncological effects (Figure 6A). To demonstrate the involvement of CDK13 in downstream effects, we used the positive EGFR pathway inhibitor drug, cetuximab, and the CDK12 and CDK13 specific inhibitor, SR-4835. We found that the activation of the EGFR pathway was higher in H1299 cells overexpressing CCNO, and the activation of the pathway was reduced after inhibition of CDK12 and CDK13. When cetuximab and SR-4835 were combined (S + C), H1299 cells with a high expression of CCNO were more sensitive to its response, increasing the sensitivity of tumor cells to chemotherapeutic drugs. We performed IHC staining of tumor tissues in correspondingly treated nude mice and found that the staining intensity of P-EGFR was consistent with that found in western blot (Figure 6B).
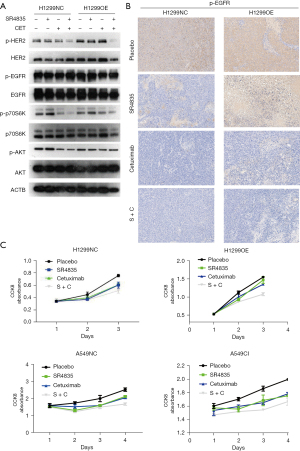
Also, we obtained the same results for the H1299, H1299OE, A549 cells, and A549CI cell lines with the same treatment of control, cetuximab, SR-4835, and S + C, respectively (Figure 6C), and observed the amount of cell activity after 3 days to detect the proliferation rate of tumor cells.
CCNO promoted tumor cell proliferation and survival in vivo via CDK13
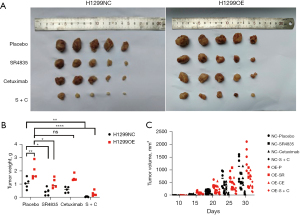
After verifying the role of CCNO and CDK13 in vitro, we explored their role in vivo. An engrafted tumor mouse model was established by subcutaneously implanting the H1299 and H1299OE cells in nude mice (Figure 7A). After the appearance of subcutaneous tumors, the nude mice were treated with either control (saline injection), positive drug cetuximab intraperitoneal injection, SR-4835 intraperitoneal injection, or a combined cetuximab and SR-4835 injection (twice weekly) and the change of tumor volume was recorded every 5 days. The tumor weights and volume of the nude mice were measured as shown (Figure 7B,7C). Statistically, we found that tumors appeared earlier and were larger in the H1299OE group. The efficacy of cetuximab in the H1299OE group of nude mice was weaker than that in the normal group, but the combination of SR-4835 and cetuximab caused tumors with a high expression of CCNO to be more sensitive to the drug and reversed their drug-resistance to cetuximab.
CDK13 was found to be a key factor in the oncological effect of CCNO
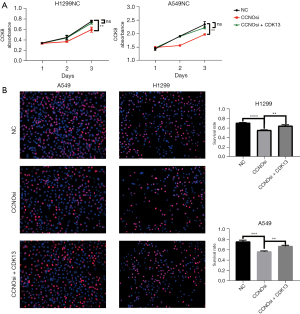
Both CCNO and CDK13 have oncological effects, and CDK13 is required for CCNO to function. In H1299 and A549 cells, we knocked down CCNO and overexpressed CDK13 on top of knockdown CCNO, respectively, and we found that both the CCK8 assay and EdU cell staining showed the same result (Figure 8A,8B): cell activity decreased, and cell proliferation was reduced after CCNO knockdown. Overexpression of CDK13 reversed this phenomenon, thus demonstrating that CDK13 is a key factor in the oncological effect of CCNO.
Discussion
The present study provides additional data demonstrating the significance of CCNO in lung cancer development. In terms of the mechanisms involved, we found that CCNO interacts with CDK13, resulting in increased EGFR signaling and cetuximab tolerance. CDK13 inhibitors reduced EGFR signaling and sensitized cetuximab for the treatment of lung cancer. This provides evidence for the precise selection of therapeutic drugs for LUAD treatment.
CCNO was initially identified as a cyclin-like uracil DNA glycosylase. However, its expression in LUAD has remained unclear. Therefore, we investigated CCNO expression in lung cancer. Using transcriptomics, we determined that the CCNO gene was highly expressed in LUAD, and data from the TCGA and GEO databases suggested that CCNO expression was much greater in tumors than in normal tissues. Following this, we evaluated the protein expression level of CCNO and demonstrated that CCNO expression was higher in lung cancer tissues than in paraneoplastic tissues. Due to the fast proliferation of tumor cells, cyclin is often expressed at a greater level in tumors, leading to a breakdown of cell cycle control. Consequently, we verified that CCNO might be employed as a diagnostic and prognostic marker for lung cancer.
To further explore the influence of CCNO on the development of LUAD, we examined whether CCNO enhances lung cancer proliferation. We evaluated the baseline expression in lung cancer cells and chose A549 and H1299 cells for the analysis. We found that downregulation of CCNO expression in LUAD cells significantly inhibited tumor cell growth. We examined the potential of CCNO to increase the proliferation of individual cells using clonogenic tests, and these results showed that aberrant CCNO expression promoted lung carcinogenesis. It has also been observed that cyclin can govern cell invasion (17,18), while CCNO has not been known to have this ability. Downregulation of CCNO expression greatly decreased LUAD cells’ invasive metastatic potential. Since the most fundamental role of cyclins is to bind CDKs to drive the cell cycle and hence govern cell proliferation, we investigated the link between CCNO expression and the lung cancer cell cycle. The G0-G1 phase of the cell cycle was inhibited by CCNO downregulation. It has been reported that CCNO acts by binding to CDK2, which is the CDK that governs the cellular G1-S phase transition (19); hence, the foregoing findings show that CCNO regulates LUAD cell proliferation, invasion, and metastasis by binding to cycle-regulated CDKs.
In addition to binding to CDK2 to control the cell cycle, CCNO is crucial to the formation of lymphocytes, DNA damage repair, and apoptosis (7,20,21). We studied whether CCNO promotes LUAD development via unique pathways. Through bioinformatics analysis, we discovered CCNO-regulated genes grouped inside conventional cell cycle regulatory networks and identified previously described CCNO-related signaling pathways, such as the apoptosis route and the redox pathway. Surprisingly, we discovered the PI3K-AKT signaling pathway in the clustering of signaling pathways. The AKT pathway is associated with increased cancer cell proliferation and survival (22). Consequently, CCNO has other biological activities beyond cell cycle control. To investigate the unique roles of CCNO, we identified its interacting proteins using IP-MS. As is well known, the classic mode of action of cyclin is to bind to cyclin kinase in a dimeric form, activating downstream molecular signaling pathways through phosphorylation. A series of molecules found in the IP-MS, unexpectedly, we discovered that the CDK component involved in transcriptional regulation (23), CDK13, interacts with CCNO. CDK13, unlike cell cycle-associated CDKs, primarily regulates RNA transcription and gene expression by binding to cyclin K (24). Our findings indicate that CCNO is a new CDK13-interacting protein and that CDK13 is co-expressed with CCNO. To demonstrate that CCNO operates via CDK13, we observed that inhibition of CDK13 reversed the proliferation of cells induced by overexpression of CCNO. We accomplished this by silencing CDK13 expression and treating cells with the CDK13 inhibitor SR-4835. Thus, CCNO serves as a regulator by binding to CDK13.
We hypothesized that since CCNO controls genes involved in the AKT signaling cascade and CDK12, a member of the same family as CDK13, may directly phosphorylate EGFR and HER2 and so boost downstream signaling, CCNO may promote EGFR signaling by binding to CDK13. Overexpression of CCNO enhanced EGFR and HER2 signaling considerably and was not blocked by the EGFR monoclonal antibody cetuximab, indicating that CCNO may activate EGFR independently of EGF. Nevertheless, the CDK13 inhibitor SR-4835 effectively suppressed CCNO overexpression-mediated EGFR activation, indicating that CCNO is a conventional therapeutic target for LUAD, although cetuximab is unsuccessful (25). Our results suggest that combining cetuximab and SR-4835 suppresses tumor cells more effectively than either treatment alone. In addition, we conducted in vivo tests to demonstrate that SR-4835 can enhance the effectiveness of cetuximab. These findings imply that elevated expression of CCNO in LUAD drives natural resistance to cetuximab by stimulating the EGFR signaling pathway via CDK13 and that suppression of the CCNO-CDK13 complex might enhance cetuximab’s effectiveness.
Conclusions
In this study, we discovered that CCNO may serve as a new diagnostic and prognostic biomarker for the development of LUAD. The expression level of CCNO has a statistically significant impact on the T stage, N stage, TNM stage, pathological stage, and prognosis of LUAD. And a new precision chemotherapy regimen is provided to enhance the efficacy of cetuximab, thereby establishing a new strategy (26) for future drug development and selection.
Acknowledgments
Funding: This study was supported by the Major Science and Technology Innovation Project of Shandong Province (No. 2020CXGC011303 to Hui Tian), the Special Fund for Taishan Scholar Program of Shandong Province (No. ts201712087 to Hui Tian), and the Clinical Research Project of Minimally Invasive Esophagectomy Based on Enhanced Rehabilitation Surgical Technology (No. 2020SDUCRCA013 to Hui Tian), The National Natural Science Foundation of China (Nos. 81803096, 81972863, 81627901, and 82030082 to Hui Tian).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-437/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-437/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-437/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-437/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for this study was granted by the Medical Ethics Committee of Qilu Hospital of Shandong University (No. KYLL-2019(KS)-046). Informed consent was obtained from each patient or their legal guardians. All animal experiments were approved by the Medical Ethics Committee of Shandong University (No. KYLL-2021(KS)-1053), in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Arellano M, Moreno S. Regulation of CDK/cyclin complexes during the cell cycle. Int J Biochem Cell Biol 1997;29:559-73. [Crossref] [PubMed]
- Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) 2017;9:52. [Crossref] [PubMed]
- Muller SJ, Caradonna S. Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim Biophys Acta 1991;1088:197-207. [Crossref] [PubMed]
- Hagen L, Kavli B, Sousa MM, et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J 2008;27:51-61. [Crossref] [PubMed]
- Roig MB, Roset R, Ortet L, et al. Identification of a novel cyclin required for the intrinsic apoptosis pathway in lymphoid cells. Cell Death Differ 2009;16:230-43. [Crossref] [PubMed]
- Ma JY, Ou-Yang YC, Luo YB, et al. Cyclin O regulates germinal vesicle breakdown in mouse oocytes. Biol Reprod 2013;88:110. [Crossref] [PubMed]
- Li L, Cao Y, Zhou H, et al. Knockdown of CCNO decreases the tumorigenicity of gastric cancer by inducing apoptosis. Onco Targets Ther 2018;11:7471-81. [Crossref] [PubMed]
- Wang J, Chen S. RACK1 promotes miR-302b/c/d-3p expression and inhibits CCNO expression to induce cell apoptosis in cervical squamous cell carcinoma. Cancer Cell Int 2020;20:385. [Crossref] [PubMed]
- Chou J, Quigley DA, Robinson TM, et al. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov 2020;10:351-70. [Crossref] [PubMed]
- Ansari D, Andersson R, Bauden MP, et al. Protein deep sequencing applied to biobank samples from patients with pancreatic cancer. J Cancer Res Clin Oncol 2015;141:369-80. [Crossref] [PubMed]
- Cheng L, Zhou S, Zhou S, et al. Dual Inhibition of CDK12/CDK13 Targets Both Tumor and Immune Cells in Ovarian Cancer. Cancer Res 2022;82:3588-602. [Crossref] [PubMed]
- Qi JC, Yang Z, Lin T, et al. CDK13 upregulation-induced formation of the positive feedback loop among circCDK13, miR-212-5p/miR-449a and E2F5 contributes to prostate carcinogenesis. J Exp Clin Cancer Res 2021;40:2. [Crossref] [PubMed]
- Choi HJ, Jin S, Cho H, et al. CDK12 drives breast tumor initiation and trastuzumab resistance via WNT and IRS1-ErbB-PI3K signaling. EMBO Rep 2019;20:e48058. [Crossref] [PubMed]
- Tadesse S, Duckett DR, Monastyrskyi A. The promise and current status of CDK12/13 inhibition for the treatment of cancer. Future Med Chem 2021;13:117-41. [Crossref] [PubMed]
- Montalto FI, De Amicis F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020;9:2648. [Crossref] [PubMed]
- Loukil A, Cheung CT, Bendris N, et al. Cyclin A2: At the crossroads of cell cycle and cell invasion. World J Biol Chem 2015;6:346-50. [Crossref] [PubMed]
- Hume S, Dianov GL, Ramadan K. A unified model for the G1/S cell cycle transition. Nucleic Acids Res 2020;48:12483-501. [Crossref] [PubMed]
- Villa M, Gialitakis M, Tolaini M, et al. Aryl hydrocarbon receptor is required for optimal B-cell proliferation. EMBO J 2017;36:116-28. [Crossref] [PubMed]
- Sharbeen G, Yee CW, Smith AL, et al. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J Exp Med 2012;209:965-74. [Crossref] [PubMed]
- Song M, Bode AM, Dong Z, et al. AKT as a Therapeutic Target for Cancer. Cancer Res 2019;79:1019-31. [Crossref] [PubMed]
- Wu C, Xie T, Guo Y, et al. CDK13 phosphorylates the translation machinery and promotes tumorigenic protein synthesis. Oncogene 2023;42:1321-30. [Crossref] [PubMed]
- Greenleaf AL. Human CDK12 and CDK13, multi-tasking CTD kinases for the new millenium. Transcription 2019;10:91-110. [Crossref] [PubMed]
- Mazzarella L, Guida A, Curigliano G. Cetuximab for treating non-small cell lung cancer. Expert Opin Biol Ther 2018;18:483-93. [Crossref] [PubMed]
- Lei P, Zhang J, Liao P, et al. Current progress and novel strategies that target CDK12 for drug discovery. Eur J Med Chem 2022;240:114603. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


