
Prediction of clinicopathological features, multi-omics events and prognosis based on digital pathology and deep learning in HR+/HER2− breast cancer
Highlight box
Key findings
• Our study quickly identified effective therapeutic targets for the precision treatment of HR+/HER2− breast cancer based on WSIs using deep-learning.
What is known and what is new?
• Our approach has high accuracy for predicting histological grade, Ki67, TP53 mutation status, G2-M checkpoint pathway, the levels of iTILs and sTILs, CD8A and PDCD1 mRNA expression.
What is the implication, and what should change now?
• Our research has high practical value in quickly identifying effective therapeutic targets for precise treatment and predicting prognosis.
Introduction
Breast cancer has the highest morbidity and mortality among women in the world (1,2). In current clinical practice, breast cancer is classified into molecular subtypes including luminal A, luminal B, HER2-enriched and triple-negative breast cancer (TNBC) (3,4). Hormone receptor (HR)+/human epidermal growth factor receptor 2 (HER2)− breast cancer refers to breast cancer that expresses the estrogen receptor (ER) or the progesterone receptor (PR) and does not express HER2. HR+/HER2− is the most common type of breast cancer, accounting for 50–79% of cases (5,6). Although a number of therapies, including endocrine therapy, have improved the prognosis of HR+/HER2− breast cancer, there are still clinical problems such as long-term recurrence (7,8). Therefore, precision treatment with targeted therapies is important for HR+/HER2− breast cancer. Although there are traditional laboratory methods for the detection of treatment targets, these methods are expensive, slow and inconvenient, limiting their broad use in clinical practice.
In recent years, deep learning has been widely used in cancer imaging research, especially for predicting some targets related to precise treatment and patient prognosis (9-11). The theoretical basis of its prediction based on deep learning is that computers can mine and learn the mapping relationship between pathological morphology and molecular features and form a neural network model to realize molecular feature prediction based on whole-slide images (WSIs). Previous studies have attempted to predict the molecular characteristics and prognosis of non-small cell lung cancer, gastrointestinal tumors, pan-cancer, and bladder cancer (12-16). In breast cancer, previous studies have tried to predict lymph node status, improve histological grade, and determine hormonal receptor status (17-21). However, there have been few studies on the prediction of therapeutic targets and prognoses of HR+/HER2− breast cancer based on digital pathology.
First, we constructed a cohort of 421 HR+/HER2− breast cancer patients with multi-omics data and pathological WSIs. Second, we set up an analysis workflow including image preprocessing, tissue type classification, molecular feature prediction and prognosis prediction. Third, we predicted clinicopathological features, gene mutations, gene set enrichment analysis (GSEA) pathways, immunotherapy markers, and prognosis based on the WSIs. We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-445/rc).
Methods
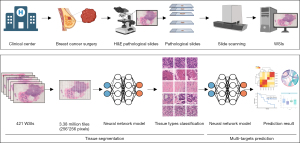
Prediction workflow based on deep learning
The workflow of our study is shown in Figure 1. First, we collected hematoxylin and eosin (H&E)-stained histological slides from patients after surgery. We used a NanoZoomer digital pathology scanner at ×40 to scan H&E-stained histological slides and generated digital WSIs. Then, WSIs were cut into image tiles, and background image tiles were filtered out. The deep learning-based analysis pipeline consisted of two convolutional neural networks (CNNs) in series. The first was a tile-level tissue type classifier which was developed in our previous study (21). The second CNN was trained based on the tiles of certain tissue type to predict clinicopathological features, somatic mutations, important cancer-related pathways, immunotherapy biomarkers and prognosis. We selected common clinicopathological features for prediction, including pathological T category, pathological N category, histological grade, and Ki67. Histological grades were assigned according to the World Health Organization (WHO) histological grade standard (22). We used 15% as the cut-off value for Ki67 (23): expression levels higher than 15% were defined as high Ki67, while expression levels lower than 15% were defined as low Ki67. In addition, we predicted somatic mutations with a mutation frequency greater than 4%. What’s more, we selected six cancer related pathways from the Molecular Signatures Database (MSigDB) and attempted to predict whether cancer related pathways are activated based on the ssGSEA score. Last but not least, we attempted to predict key immune-related targets related to immunotherapy.
Patients and dataset
We retrospectively collected H&E-stained pathological slides from HR+/HER2− breast cancer patients after radical mastectomy or breast conserving surgery between January 2013 and December 2014 at FUSCC and scanned them to generate WSIs according to standard protocols. The inclusion criteria for this cohort study were as follows: (I) the diagnosis of HR+/HER2− breast cancer was confirmed by histopathology or cytology; and (II) there was no evidence of distant metastasis. The exclusion criteria were as follows: (I) no formalin-fixed, paraffin-embedded (FFPE) samples were available; (II) patients with slides or WSIs of poor quality (large artifacts, debris, pen marks or blurred images) (15); and (III) patients lost to follow-up (Figure S1). Two pathologists independently conducted the quality control of all WSIs. Disagreements on the quality control results were discussed and resolved through negotiation. Only those patients with high-quality WSIs were included in the study. Finally, a total of 421 patients were selected.
Data preprocessing
Background image region exclusion
To reduce the computational burden and shorten the training time, we used the rectangle tool of ImageScope software to generate one or two region of interests (ROIs) that included the vast majority of the invasive breast cancer areas and excluded the dragged tissue and background areas.
Digital pathological image tiling
WSIs were cut into image tiles before being fed into CNNs for modelling. We used the OpenSlide library of the MATLAB software to divide each WSI into 256×256 pixel square image tiles. The white background part in the ROI area were filtered out during MATLAB image tiling. Image tiles with limited tissues (defined as more than half of the pixels within the tile were >210) were also discarded (21). Four hundred and twenty-one slides were cut into 3,388,890 image tiles (Figure S2). Tiles were stored in PNG format, and the information about the patient ID was contained in the filenames.
Tissue type segmentation
A WSI contains a variety of tissue types, including tumor, stroma, immune infiltration, normal duct, and necrosis or hemorrhage. We selected certain tissue types of image tiles for model development. For example, we used tumor image tiles to predict somatic mutations and used tumor, stroma and immune infiltrates tiles to predict immunotherapy biomarkers such as CD274 and PDCD1. Therefore, we used a tile tissue type classifier we previously developed to realize the automatic classification of certain types of image tiles (21). We used the ResNet-18 as our tissue type segmentation CNN architecture, which is a commonly used residual neural network (24). The other hyperparameters were set as follows: cross-entropy loss as the loss function and ADAM algorithm for optimization. We took 256 image tiles in the training set for each training epoch and trained the model for 200 epochs. The learning rate was set to 0.001 and the momentum was set to 0.9.
Image-based identification of clinicopathological features and multi-omics molecular features using deep learning
Clinicopathological features and multi-omics molecular labels
The baseline clinical and pathological characteristics of the HR+/HER2− breast cancer patient cohort are detailed in Table 1.
Table 1
| Variables | HR+/HER2− breast cancer cohort (N=421) |
|---|---|
| Age, years | |
| ≤50 | 173 (41.1) |
| >50 | 248 (58.9) |
| T category | |
| T1 | 199 (47.3) |
| T2 | 218 (51.8) |
| T3 | 3 (0.7) |
| NA | 1 (0.2) |
| N category | |
| N0 | 190 (45.1) |
| N1 | 134 (31.9) |
| N2 | 59 (14.0) |
| N3 | 38 (9.0) |
| Tumor grade | |
| I–II | 284 (67.5) |
| III | 117 (27.8) |
| Unknown | 20 (4.8) |
| Surgery type | |
| Non-BCS | 417 (99.0) |
| BCS | 4 (1.0) |
| Radiotherapy | |
| No | 253 (60.1) |
| Yes | 128 (30.4) |
| Unknown | 40 (9.5) |
| Chemotherapy | |
| No | 86 (20.4) |
| Yes | 312 (74.1) |
| Unknown | 23 (5.5) |
| Endocrine therapy | |
| No | 11 (2.61) |
| Yes | 359 (85.27) |
| Unknown | 51 (12.12) |
Data are presented as number (percentage) of patients. BCS, breast conserving surgery.
First, we aimed to predict the clinicopathological features of the HR+/HER2− breast cancer cohort, including T category, N category, histological grade, and Ki67. We set corresponding clinicopathological feature labels for patients according to their clinical data. Second, we developed models to predict somatic mutations with a frequency of ≥4% from WSIs. Patients with the corresponding gene mutations were labelled as the “positive” cases. Third, we explored the prediction of biological pathways. We selected six breast cancer-related hallmark gene sets from MSigDB including the G2-M checkpoint pathway, DNA repair pathway, PI3K/AKT/mTOR signalling pathway, IFN-γ response pathway, angiogenesis pathway and early estrogen response pathway (25). For each of them, we calculated an enrichment score for each sample using the single sample GSEA method to measure the relative activity of the corresponding biological pathways (26). Patients whose ssGSEA scores were higher than the median were labelled as the “positive” cases. Fourth, we attempted to predict key immunotherapeutic biomarkers, including stromal tumor-infiltrating lymphocytes (sTILs), intratumoral tumor-infiltrating lymphocytes (iTILs), PD-L1 expression, PD-1 expression, and CD8 expression. iTILs were defined as lymphocytes within nests of carcinoma having cell-to-cell contact with no intervening stroma, while sTILs were defined as lymphocytes located in the stroma between the tumor nests. TILs were evaluated according to the 2015 TILs evaluation recommendation by the International TILs Working Group (27). TILs scores were independently assessed according to the recommendations by two experienced pathologists who were unaware of the clinical information of the patients. Disagreements between the two raters were resolved through discussion and consensus. The expression levels of PD-L1 and PD-1 were measured by CD274 and PDCD1 mRNA expression, respectively (28), and the expression level of CD8 was measured by CD8A mRNA expression (29). Patients whose mRNA expression was higher than the median were labelled as the “positive” cases.
Division strategy for the training set, verification set and test set
We used the hold-out method to train and validate CNN models predicting clinicopathological features and multi-omics molecular features. The hold-out method is one of the most common methods used to evaluate the performance of a machine learning model. We divided the training set, the validation set and the test set into 3:1:1. Stratified random sampling reduces the sampling error to avoid the impact on the results due to data bias. We tessellated the WSIs into tiles as described previously. All of the tiles inherited the labels of the corresponding patients.
Training of the neural network and visualization of prediction results
CNN models were trained on the tiles from the training set, validated on the tiles from the verification set, and tested on the tiles from the test set. For the clinicopathological feature and multi-omics feature prediction, we used the ResNet-18 as the CNN architecture (24). The other parameters were set as follows: cross-entropy loss as loss function; the learning rate was set to 0.001; the batch size was set to 256 and the ADAM algorithm for optimization.
Tiles in the validation set and test set acquired a prediction score output from the model. The prediction score was calculated by averaging all the tile scores from the corresponding patient. The patient-level prediction scores were used for receiver operating characteristic (ROC) analysis. A model was saved after each training epoch, and the model with the highest area under the curve (AUC) in the validation set was considered the best model and further evaluated in the test set.
Two visualization methods were used to display the predicted results of the CNN models and identify pathological patterns associated with the clinicopathological and molecular characteristics. First, for each prediction task, the CNN model outputs a score for each tile. We visually described the morphological characteristics of tiles with the highest prediction scores in true positive patients. Second, class activation maps were established for these tiles (30). This approach enabled identification of the local regions most relevant to the prediction target.
Prediction of prognosis using deep learning
DeepSurv neural network
DeepSurv is a neural network based on the Cox proportional risk model to model the relationship between patients’ covariates and prognosis. The DeepSurv architecture consists of fully connected layers (20, 128, 512, 2,048, 1,024, 256, 64, and 8 neurons) followed by dropout layers and ReLU activation. Patients’ or tiles’ features were took as input and the model output a risk score which estimated the log-risk function in the Cox model. The Cox loss function was used as the loss function and the ADAM algorithm was used for optimization. For the other hyperparameters settings, the dropout rate was set to 0.2; the learning rate was set to 0.001; momentum was set to 0.9; and the model was trained for 100 epochs.
Modelling strategies
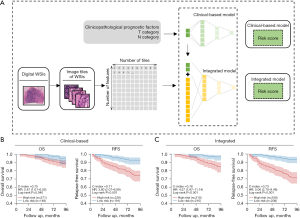
We compared two modelling strategies. The first was a clinic-based model. We selected statistically significant variables according to univariate Cox analysis to determine the clinical prognostic factors. The second modelling strategy is an integrated model. We selected 500 tiles of all five tissue types for each patient and fed them into a neural network for deep feature extraction. Deep features of image tiles and clinical prognostic features were combined to build the integrated model to predict overall survival (OS) and relapse-free survival (RFS).
Three-fold cross validation
We used a three-fold cross-validation strategy to train and validate the prognostic models (15,21). The entire cohort was divided into three parts of equal size. One part was used as the validation set, and the other two parts were used as the training set. A model was saved after each training epoch, C-index value was used to evaluate the prediction accuracy and the best model was selected according to the maximum value of the C-index in the validation set (31). This method was repeated three times until each part had been used as the validation set. Each of the three best models output risk scores for their corresponding validation set. These scores were concatenated and therefore each patient obtained a final risk score. The median value of the risk scores was used to separate high-risk and low-risk patients and hazard ratio was calculated between groups.
Statistical analysis
PyTorch framework was used for deep learning experiments (32). The AUC, along with its 95% confidence interval (CI), was used to evaluate the prediction accuracy. RFS was defined as the time from diagnosis to first recurrence, diagnosis of contralateral breast cancer, or death from any cause. OS was defined as the time from randomization to death due to any cause. Survival curves were drawn using the Kaplan-Meier method, and survival differences between groups were compared using the log-rank test and the Cox proportional hazards (CPH) model. The C-index was used to evaluate the prediction accuracy of prognostic models.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Fudan University Shanghai Cancer Center (FUSCC) Ethics Committee (approval No. 050432-4-1911D). All patients provided written informed consent.
Results
Establishment of a HR+/HER2− breast cancer cohort
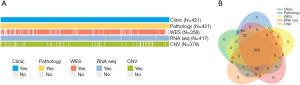
We retrospectively analyzed 421 HR+/HER2− breast cancer patients between January 2013 and December 2014 at FUSCC. Among these 421 patients, all patients had clinical and pathology data, 358 had whole-exome sequencing (WES) data, 417 had RNA sequencing data, and 379 had copy number variation (CNV) data (Figure 2A). The Venn diagram of the HR+/HER2− breast cancer cohort showed that 323 patients had clinical data, digital pathological image data, WES data, RNA sequencing data and CNV data available simultaneously (Figure 2B). Clinicopathological characteristics of the cohort was shown in Table 1.
Classification of tissue types using CNN
Using the tissue type classifier developed in our previous study, we classified all image tiles into five tissue types: tumor, stroma, immune infiltrates, normal duct, and necrosis (21). Tumor, stroma, immune infiltrate, normal duct, and necrosis image tiles accounted for 37.92%, 53.43%, 2.42%, 3.59% and 2.63%, respectively (Figure 3A). Representative tiles for each of the five tissue types are shown in Figure 3B. We present four representative examples of tissue type classification results in Figure 3C. For the following prediction tasks, we selected certain tissue types of tiles that were related to the prediction targets (Table 2).

Table 2
| Prediction categories | Prediction targets | Tissue types of tiles sampled |
|---|---|---|
| Clinical pathological features | T1 | Tumor |
| T2–3 | Tumor | |
| N0 | Tumor | |
| N1–3 | Tumor | |
| Grade I | Tumor | |
| Grade II | Tumor | |
| Grade III | Tumor | |
| Low Ki67 | Tumor | |
| High Ki67 | Tumor | |
| Somatic mutations | PIK3CA | Tumor |
| TP53 | Tumor | |
| GATA3 | Tumor | |
| MAP3K1 | Tumor | |
| KMT2C | Tumor | |
| AKT1 | Tumor | |
| PTEN | Tumor | |
| FAT3 | Tumor | |
| SF3B1 | Tumor | |
| Gene set enrichment analysis scores | Angiogenesis | Tumor |
| DNA repair | Tumor | |
| Estrogen response early | Tumor | |
| G2-M checkpoint | Tumor | |
| IFN-γ response | Tumor | |
| PI3K/AKT/mTOR signaling | Tumor | |
| Immunotherapy biomarkers | iTILs | Tumor |
| sTILs | Stroma and immune infiltrates | |
| CD8A mRNA | Stroma and immune infiltrates | |
| PDCD1 mRNA | Stroma and immune infiltrates | |
| CD274 mRNA | Tumor, stroma and immune infiltrates | |
| Prognosis | OS | All five tissue types |
| DFS | All five tissue types |
iTILs, intratumoral tumor-infiltrating lymphocytes; sTILs, stromal tumor-infiltrating lymphocytes; OS, overall survival; DFS, disease-free survival.
Prediction of clinicopathological features
We found that histological grade and Ki67 had high prediction accuracy. In the test set, grade I, grade II and grade III can be predicted with AUCs of 0.68 (95% CI: 0.46–0.89), 0.82 (95% CI: 0.72–0.92) and 0.90 (95% CI: 0.84–0.97), respectively; low Ki67 and high Ki67 were predicted with AUCs of 0.81 (95% CI: 0.69–0.92) and 0.80 (95% CI: 0.66–0.94), respectively. However, the models for predicting T category and pathological N category did not achieve perfect accuracy (Figure 4, Table 3).

Table 3
| Clinicopathological features | Validation AUC (95% CI) | Test AUC (95% CI) |
|---|---|---|
| T1 | 0.66 (0.54–0.78) | 0.57 (0.44–0.70) |
| T2–3 | 0.61 (0.49–0.74) | 0.54 (0.41–0.67) |
| N0 | 0.61 (0.48–0.74) | 0.51 (0.39–0.64) |
| N1–3 | 0.59 (0.47–0.72) | 0.52 (0.39–0.65) |
| Grade I | 0.97 (0.92–1.00) | 0.68 (0.46–0.89) |
| Grade II | 0.81 (0.71–0.91) | 0.82 (0.72–0.92) |
| Grade III | 0.80 (0.70–0.89) | 0.90 (0.84–0.97) |
| Low Ki67 | 0.85 (0.71–0.99) | 0.81 (0.69–0.92) |
| High Ki67 | 0.83 (0.70–0.96) | 0.80 (0.66–0.94) |
AUC, area under the curve; CI, confidence interval.
We investigated the morphological patterns that were associated with the clinicopathologic characteristics. For a certain clinicopathological feature, we visually examined the tiles with the highest prediction score from the true positive patients. Tiles indicating grade I had the morphologic features of little cellular atypia and rare mitosis, and obvious glandular tubes. Tiles indicating grade II had more cellular atypia, more mitosis and less glandular tube than grade I. Tiles indicating grade III had features of conspicuously pleomorphic cells, frequent mitoses and no glandular tube (Figure S3A-S3C). The characteristics of high Ki67 image tiles were similar to those of grade III tiles, with lymphocyte infiltration. In addition, low Ki67 image tiles were similar to those of grade I–II with rare lymphocyte infiltration (Figure S3D,S3E).
Prediction of multi-omics molecular features
In this section, we aimed to develop CNN models for predicting multi-omics molecular features from WSIs. We first predicted somatic mutations with a frequency of ≥4%; the AUCs ranged from 0.50 to 0.85 for the validation set and from 0.42 to 0.68 for the test set (Figure 5, Table 4). Specifically, TP53 mutation and GATA3 mutation can be predicted with AUCs of 0.68 (95% CI: 0.56–0.81) and 0.68 (95% CI: 0.47–0.89), respectively in the test set. Second, we predicted the GSEA scores of six cancer-related pathways. The AUCs ranged from 0.63 to 0.87 for the validation set and from 0.48 to 0.79 for the test set (Figure 5, Table 4). Third, we attempted to predict several key biomarkers associated with immunotherapy responses, including CD274 mRNA expression, CD8A mRNA expression, PDCD1 mRNA expression, sTILs and iTILs. The AUCs ranged from 0.59 to 0.76 for the validation set and from 0.51 to 0.78 for the test set (Figure 5, Table 4).
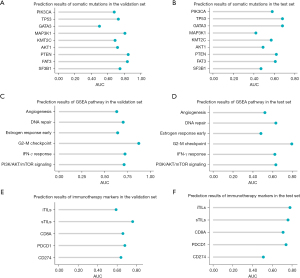
Table 4
| Prediction categories | Multi-omics molecular features | Validation AUC (95% CI) |
Test AUC (95% CI) |
|---|---|---|---|
| Somatic mutations | PIK3CA | 0.68 (0.56–0.80) | 0.58 (0.44–0.71) |
| TP53 | 0.73 (0.62–0.85) | 0.68 (0.56–0.81) | |
| GATA3 | 0.50 (0.31–0.68) | 0.68 (0.47–0.89) | |
| MAP3K1 | 0.81 (0.63–0.99) | 0.42 (0.22–0.61) | |
| KMT2C | 0.69 (0.49–0.89) | 0.57 (0.36–0.79) | |
| AKT1 | 0.72 (0.49–0.94) | 0.49 (0.27–0.71) | |
| PTEN | 0.85 (0.72–0.97) | 0.62 (0.40–0.83) | |
| FAT3 | 0.84 (0.67–1.00) | 0.61 (0.16–1.00) | |
| SF3B1 | 0.75 (0.49–1.00) | 0.47 (0.00–1.00) | |
| Gene set enrichment analysis scores | Angiogenesis | 0.63 (0.51–0.75) | 0.52 (0.39–0.65) |
| DNA repair | 0.70 (0.59–0.82) | 0.63 (0.50–0.75) | |
| Estrogen response early | 0.64 (0.50–0.77) | 0.48 (0.34–0.62) | |
| G2-M checkpoint | 0.87 (0.80–0.95) | 0.79 (0.69–0.90) | |
| IFN-γ response | 0.72 (0.61–0.84) | 0.62 (0.50–0.75) | |
| PI3K/AKT/mTOR signaling | 0.71 (0.60–0.82) | 0.63 (0.50–0.76) | |
| Immunotherapy biomarkers | iTILs | 0.59 (0.35–0.82) | 0.78 (0.55–1.00) |
| sTILs | 0.76 (0.65–0.87) | 0.76 (0.65–0.87) | |
| CD8A mRNA | 0.66 (0.54–0.77) | 0.71 (0.60–0.82) | |
| PDCD1 mRNA | 0.68 (0.56–0.79) | 0.74 (0.63–0.85) | |
| CD274 mRNA | 0.64 (0.52–0.76) | 0.51 (0.39–0.64) |
AUC, area under the curve; CI, confidence interval; iTILs, intratumoral tumor-infiltrating lymphocytes; sTILs, stromal tumor-infiltrating lymphocytes.
We displayed the representative tiles indicating certain molecular characteristics (Figure S4). Tiles indicating somatic TP53 mutation were morphologically characterized by mitosis can be seen, with necroses appeared and rare lymphocyte infiltration; tiles indicating somatic PTEN mutation were characterized by rare mitosis, rare lymphocyte infiltration, and no obvious fibrous hyperplasia; and tiles indicating somatic GATA3 mutation were characterized by rare mitosis, little/moderate cellular atypia, and no glandular tube (Figure S4A-S4C). In terms of the image tiles characteristics of cancer- related pathways, we observed cells size were similar, and with lymphocyte infiltration in DNA repair image tiles; G2-M checkpoint image tiles were characterized by many mitosis; IFN-γ response pathway image tiles were characterized by mitosis and glandular tube observed; and PI3K/AKT/mTOR signaling pathway image tiles were characterized by the polymorphic tumor cells, frequent mitoses, and high immune infiltration (Figure S4D-S4G). In addition, tiles indicating high CD8A mRNA expression had a high density of lymphocytes (Figure S4H). Moreover, those indicating high PDCD1 mRNA expression were characterized by myofibroblast hyperplasia (Figure S4I). Finally, the characteristics of the image tiles rich in iTILs were intratumoral lymphocytes can be seen, and those rich in sTILs were characterized by high density lymphocytes infiltration in stroma (Figure S4J,S4K).
Prediction of prognosis
We built clinically based prognosis prediction models based on clinical prognostic features and integrated models based on clinical prognostic features combined with deep features of image tiles. First, in the development of the clinically based models, T category, N category and radiotherapy were identified as prognostic factors by univariate analysis (Table 5). Since the negative prognostic effect of radiotherapy might be related to the selection bias, radiotherapy was not used in the subsequent modelling (33). The T category and N category were used to build a clinical-based model (Figure 6A). The clinically based models achieved a cross-validation C-index of 0.75 in the prediction of OS and 0.71 in the prediction of RFS (Figure 6B). Then, we built integrated models combining clinical features and deep features of image tiles. The integrated models achieved a cross-validation C-index of 0.76 in OS prediction and 0.73 in RFS prediction (Figure 6C). The integrated models also yielded higher HRs between the high- and low-risk groups than the clinical models.
Table 5
| Variables | Relapse-free survival | |
|---|---|---|
| HR (95% CI) | P | |
| Age, years | ||
| ≤50 | Reference | – |
| >50 | 1.09 (0.66–1.80) | 0.732 |
| T category | ||
| T1 | Reference | – |
| T2 | 1.85 (1.09–3.14) | 0.022 |
| T3 | 19.34 (5.65–66.22) | <0.001 |
| N category | ||
| N0 | Reference | – |
| N1 | 2.24 (1.10–4.53) | 0.025 |
| N2 | 4.11 (1.90–8.87) | <0.001 |
| N3 | 10.95 (5.40–22.20) | <0.001 |
| Tumor grade | ||
| I–II | Reference | – |
| III | 1.40 (0.81–2.42) | 0.230 |
| Radiotherapy | ||
| No | Reference | – |
| Yes | 2.91(1.76–4.79) | <0.001 |
| Chemotherapy | ||
| No | Reference | – |
| Yes | 1.17 (0.62–2.19) | 0.630 |
HR, hazard ratio; CI, confidence interval.
Discussion
Based on the data from the multi-omics HR+/HER2− breast cancer cohort, we collected WSIs and designed a workflow based on deep learning to train neural network models to predict the clinical and multi-omics molecular characteristics and prognosis.
In terms of predicting clinicopathological features, we found that the prediction accuracy for histological grade and Ki67 was good. The histological grade is closely related to the nature of the tumor. When the grade is higher, the prognosis of the patient is worse (22). Ki67 is an important indicator of proliferation of tumor cells, and it’s high expression is associated with poor prognosis. Studies have shown that patients with high Ki67 expression have a worse prognosis (34,35).
In predicting multi-omics molecular characteristics, we used digital pathology to predict somatic mutations, important cancer pathways and immune-related targets. For somatic mutations, the models achieved high accuracy in predicting TP53 and GATA3 mutations. Studies have shown that patients with TP53 mutations have poorer prognoses and were more likely to develop resistance to tamoxifen and aromatase inhibitors (36,37). GATA3 mutations occurred mainly in patients with luminal-like breast cancer and were associated with a favourable prognosis (38). In terms of cancer-related pathways, we achieved high accuracy in predicting the G2-M checkpoint pathway and PI3K/AKT/mTOR signalling pathway activation. Patients with tumors with G2-M checkpoint pathway activation are more likely to develop metastasis and have a worse prognosis (39). Previous studies indicated that PI3K inhibitors and AKT inhibitors improve the prognosis of patients with PI3K/AKT/mTOR pathway-activated HR+/HER2− breast cancer (40,41). Immunotherapy has excellent prospects in the treatment of breast cancer (42). Our models achieved high accuracy in predicting sTILs, iTILs, PDCD1 mRNA and CD8A mRNA expression, which were widely studied biomarkers that can distinguish patients who may benefit from immunotherapy (43-47). These results indicated that it may be possible to predict immunotherapy response based on WSI through deep learning.
Previous studies have made remarkable success in predicting patient outcomes based on WSIs using the deep learning algorithm. For example, Saillard et al. using two deep learning algorithms predicted the prognosis of liver cancer patients based on pathological sections (48). Byun et al. compared the CPH models, the random survival forest (RSF) and DeepSurv models in predicting the prognosis of renal cell carcinoma. The C-index values were 0.794, 0.789, and 0.802, respectively. The efficacy of DeepSurv was superior to the CPH models and the RSF models (49). Matsuo et al. published a study comparing the predictive efficacy of the CPH regression model based on clinicopathological features and the deep-learning neural network model in predicting prognosis in cervical cancer. The deep-learning model had higher accuracy in predicting prognosis of cervical cancer than the CPH regression model (mean absolute error, CPH regression vs. deep-learning, 43.6 vs. 30.7) (50). The above studies indicated that deep learning was a new method for effectively predicting the prognosis of different types of tumor.
Deep learning was a new, emerging neural network technique that simulates the human brain for analyzing and interpreting data. In contrast to traditional image processing methods, deep learning can extract deep features of images to predict targets. We are the first study to predict clinicopathological features, somatic mutations, important cancer-related pathways, and immune-related targets in a large-scale HR+/HER2− breast cancer cohort based on deep learning and WSIs. This research results were expected to achieve low-cost, rapid and convenient primary screening and prognostic assessment of therapeutic targets for HR+/HER2− breast cancer, and promote the clinical realization of precise diagnosis and treatment of HR+/HER2− breast cancer, which is important for selection of treatment options.
However, our study also had some limitations. First, although we developed models to predict a wide range of targets, including clinicopathological features, somatic mutations, important cancer pathways and immune-related targets, only a few targets achieved high prediction accuracy. For example, the prediction accuracy values for MAP3K1 mutation and CD274 mRNA expression need to be improved. In future studies, we may further try to use cell-level features to predict molecular events. Second, our study lacked external cohorts to validate the accuracy of the model. Further research should use The Cancer Genome Atlas (TCGA) dataset for external validation of our prediction model.
Conclusions
In conclusion, this study successfully established a deep-learning-based workflow to predict clinicopathological features, somatic mutations, important cancer pathways, immune-related targets and prognosis with HR+/HER2− breast cancer based on pathology images. Our workflow and models may promote efficient patient stratification and offers clues for artificial intelligence guided precision treatment of HR+/HER2− breast cancer.
Acknowledgments
We would like to thank all investigators who helped with the data collection and analysis. We are also grateful to all who reviewed and commented on an early draft of the paper.
Funding: This work was supported by the National Natural Science Foundation of China (grant Nos. 81572583, 81922048, 81874113, 82072922, 81903684, 92159301 and 91959207).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-445/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-445/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-445/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-445/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Fudan University Shanghai Cancer Center (FUSCC) Ethics Committee (approval No. 050432-4-1911D). All patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Veronesi U, Boyle P, Goldhirsch A, et al. Breast cancer. Lancet 2005;365:1727-41. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Prat A, Parker JS, Fan C, et al. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat 2012;135:301-6. [Crossref] [PubMed]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol 2011;5:5-23. [Crossref] [PubMed]
- Cuyún Carter G, Mohanty M, Stenger K, et al. Prognostic Factors in Hormone Receptor-Positive/Human Epidermal Growth Factor Receptor 2-Negative (HR+/HER2-) Advanced Breast Cancer: A Systematic Literature Review. Cancer Manag Res 2021;13:6537-66. [Crossref] [PubMed]
- Kay C, Martínez-Pérez C, Meehan J, et al. Current trends in the treatment of HR+/HER2+ breast cancer. Future Oncol 2021;17:1665-81. [Crossref] [PubMed]
- AlFakeeh A, Brezden-Masley C. Overcoming endocrine resistance in hormone receptor-positive breast cancer. Curr Oncol 2018;25:S18-27. [Crossref] [PubMed]
- Hanker AB, Sudhan DR, Arteaga CL. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020;37:496-513. [Crossref] [PubMed]
- Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301-9. [Crossref] [PubMed]
- Skrede OJ, De Raedt S, Kleppe A, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 2020;395:350-60. [Crossref] [PubMed]
- van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: the path to the clinic. Nat Med 2021;27:775-84. [Crossref] [PubMed]
- Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559-67. [Crossref] [PubMed]
- Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054-6. [Crossref] [PubMed]
- Courtiol P, Maussion C, Moarii M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med 2019;25:1519-25. [Crossref] [PubMed]
- Kather JN, Heij LR, Grabsch HI, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer 2020;1:789-99. [Crossref] [PubMed]
- Woerl AC, Eckstein M, Geiger J, et al. Deep Learning Predicts Molecular Subtype of Muscle-invasive Bladder Cancer from Conventional Histopathological Slides. Eur Urol 2020;78:256-64. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Naik N, Madani A, Esteva A, et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun 2020;11:5727. [Crossref] [PubMed]
- Wang Y, Acs B, Robertson S, et al. Improved breast cancer histological grading using deep learning. Ann Oncol 2022;33:89-98. [Crossref] [PubMed]
- Zheng X, Yao Z, Huang Y, et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun 2020;11:1236. [Crossref] [PubMed]
- Zhao S, Yan CY, Lv H, et al. Deep learning framework for comprehensive molecular and prognostic stratifications of triple-negative breast cancer. Fundamental Research 2022. [Epub ahead of print]. doi:
10.1016/j.fmre.2022.06.008 .10.1016/j.fmre.2022.06.008 - Rakha EA, Reis-Filho JS, Baehner F, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res 2010;12:207. [Crossref] [PubMed]
- Hashmi AA, Hashmi KA, Irfan M, et al. Ki67 index in intrinsic breast cancer subtypes and its association with prognostic parameters. BMC Res Notes 2019;12:605. [Crossref] [PubMed]
- He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: IEEE, 2016.
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015;26:259-71. [Crossref] [PubMed]
- Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol 2018;18:153-67. [Crossref] [PubMed]
- Emens LA, Molinero L, Loi S, et al. Atezolizumab and nab-Paclitaxel in Advanced Triple-Negative Breast Cancer: Biomarker Evaluation of the Impassion130 Study. J Natl Cancer Inst 2021;113:1005-16. [Crossref] [PubMed]
- Zhou B, Khosla A, Lapedriza A, et al. Learning Deep Features for Discriminative Localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: IEEE, 2016.
- Uno H, Cai T, Pencina MJ, et al. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 2011;30:1105-17. [Crossref] [PubMed]
- Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. arXiv 2019. arXiv:1912.01703.
- Yoon H, Mehta MP, Perumal K, et al. Atypical meningioma: randomized trials are required to resolve contradictory retrospective results regarding the role of adjuvant radiotherapy. J Cancer Res Ther 2015;11:59-66. [Crossref] [PubMed]
- Smith I, Robertson J, Kilburn L, et al. Long-term outcome and prognostic value of Ki67 after perioperative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): an open-label, multicentre, parallel-group, andomized, phase 3 trial. Lancet Oncol 2020;21:1443-54. [Crossref] [PubMed]
- Viale G, Regan MM, Mastropasqua MG, et al. Predictive value of tumor Ki-67 expression in two randomized trials of adjuvant chemoendocrine therapy for node-negative breast cancer. J Natl Cancer Inst 2008;100:207-12. [Crossref] [PubMed]
- Andersson J, Larsson L, Klaar S, et al. Worse survival for TP53 (p53)-mutated breast cancer patients receiving adjuvant CMF. Ann Oncol 2005;16:743-8. [Crossref] [PubMed]
- Grote I, Bartels S, Kandt L, et al. TP53 mutations are associated with primary endocrine resistance in luminal early breast cancer. Cancer Med 2021;10:8581-94. [Crossref] [PubMed]
- Jiang YZ, Yu KD, Zuo WJ, et al. GATA3 mutations define a unique subtype of luminal-like breast cancer with improved survival. Cancer 2014;120:1329-37. [Crossref] [PubMed]
- Oshi M, Takahashi H, Tokumaru Y, et al. G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer. Int J Mol Sci 2020;21:2921. [Crossref] [PubMed]
- André F, Ciruelos EM, Juric D, et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: final overall survival results from SOLAR-1. Ann Oncol 2021;32:208-17. [Crossref] [PubMed]
- Jones RH, Casbard A, Carucci M, et al. Fulvestrant plus capivasertib versus placebo after relapse or progression on an aromatase inhibitor in metastatic, oestrogen receptor-positive breast cancer (FAKTION): a multicentre, andomized, controlled, phase 2 trial. Lancet Oncol 2020;21:345-57. [Crossref] [PubMed]
- Emens LA. Breast Cancer Immunotherapy: Facts and Hopes. Clin Cancer Res 2018;24:511-20. [Crossref] [PubMed]
- Byrne A, Savas P, Sant S, et al. Tissue-resident memory T cells in breast cancer control and immunotherapy responses. Nat Rev Clin Oncol 2020;17:341-8. [Crossref] [PubMed]
- Miao Y, Wang J, Li Q, et al. Prognostic value and immunological role of PDCD1 gene in pan-cancer. Int Immunopharmacol 2020;89:107080. [Crossref] [PubMed]
- Oner G, Altintas S, Canturk Z, et al. The immunologic aspects in hormone receptor positive breast cancer. Cancer Treat Res Commun 2020;25:100207. [Crossref] [PubMed]
- Schalper KA, Velcheti V, Carvajal D, et al. In situ tumor PD-L1 Mrna expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res 2014;20:2773-82. [Crossref] [PubMed]
- Schmid P, Adams S, Rugo HS, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379:2108-21. [Crossref] [PubMed]
- Saillard C, Schmauch B, Laifa O, et al. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology 2020;72:2000-13. [Crossref] [PubMed]
- Byun SS, Heo TS, Choi JM, et al. Deep learning based prediction of prognosis in nonmetastatic clear cell renal cell carcinoma. Sci Rep 2021;11:1242. [Crossref] [PubMed]
- Matsuo K, Purushotham S, Jiang B, et al. Survival outcome prediction in cervical cancer: Cox models vs deep-learning model. Am J Obstet Gynecol 2019;220:381.e1-381.e14. [Crossref] [PubMed]


