
PCK2 inhibits lung adenocarcinoma tumor cell immune escape through oxidative stress-induced senescence as a potential therapeutic target
Highlight box
Key findings
• An increased expression of PCK2 may be employed as a novel prognostic biomarker in patients with lung adenocarcinoma and has been shown to increase OS, DSS, and PFI.
What is known and what is new?
• Transcriptional activation of PCK2 by PGC-1β and ERR-α promotes glutamine metabolism and colorectal cancer survival.
• This study investigated the impact of PCK2 on tumor cell behavior and the underlying mechanism.
What is the implication, and what should change now?
• Improving the prognosis of lung adenocarcinoma by interference with PCK2 may be possible since it induces senescence through the oxidative stress response and blocks the immune escape of tumor cells. These results point to a probable target anticancer treatment development in lung adenocarcinoma.
Introduction
Non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) account for over 95% of lung cancer diagnoses and deaths globally (1). Lung adenocarcinoma is a common form of lung cancer, accounting for roughly 40% of all lung cancer cases (2). NSCLC is further subdivided into three types: squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (2). Surgery followed by radiation treatment and chemotherapy has been shown to considerably increase the 5-year survival rate for patients with early-stage lung adenocarcinoma (with stages IA, IB, IIA, and IIB as 83%, 68%, 60%, and 53%, respectively) (3). On the other hand, individuals with advanced lung adenocarcinoma are seldom treated with the same methods, and their 5-year survival rate is approximately 10% (4). While historically, patients with lung adenocarcinoma were treated with minimally invasive surgery, radiotherapy, and chemotherapy, with the advent and widespread use of biological therapy and immunotherapy, these modalities have been largely replaced, and patient survival times have gradually increased (5). The postoperative recurrence rate and the death rate from lung cancer may be drastically decreased in patients who undergo urgent surgical therapy (6). As a result, it is crucial that we find effective target molecules in both early diagnosis and therapy as promptly as possible.
At present, the abnormal metabolism of tumors is a hot research topic, and most studies are conducted on the three basic metabolisms (7). Gluconeogenesis refers to the process in which cells use non-sugar precursors, such as glycerol, lactic acid, pyruvic acid, and sugar-producing amino acids, to generate free glucose or glycogen (8). In many tumor cells, gluconeogenesis plays a crucial regulatory function in the aerobic glycolysis metabolic pathway, although this mechanism receives far less attention than glycolysis and oxidative phosphorylation in cancer research (9,10). Despite the availability of oxygen, glycolysis continues to be an important energy source for many tumor cells, and the term “aerobic glycolysis of tumors” (11) describes this phenomenon. The gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK) catalysis the GTP-dependent oxaloacetate (OAA) to phosphoenolpyruvate (PEP) reaction. The two versions of this enzyme are PCK1 (PEPCK-C) in the cytoplasm and phosphoenolpyruvate carboxykinase 2 (PCK2) (PEPCK-M) in the mitochondria (12). Recent research has shown the significance of PCK2 in biological processes including the emergence and progression of many cancers (13). Renal cell carcinoma development may be slowed by increasing endoplasmic reticulum stress and the responsiveness to the drug sunitinib when genes normally silenced by epigenetic mechanisms are activated. Transcriptional activation of PCK2 by PGC-1β and ERR-α promotes glutamine metabolism and colorectal cancer survival (14), while down-regulation of PCK2 in melanoma tumor-regenerating cells modifies the tricarboxylic acid cycle (15). However, research on the function of PCK2 in lung cancer is limited.
In this study, we primarily analysed several datasets, including The Cancer Genome Atlas (TCGA), to determine how often PCK2 was expressed and how well it predicted survival for patients with lung adenocarcinoma. To further investigate the impact of PCK2 on tumor cell behavior and the underlying mechanism, we conducted bioinformatics analysis and validation. We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-542/rc).
Methods
Methods for data mining in the TCGA database
We investigated PCK2 expression in TCGA pan-cancer samples and in lung cancer and adjacent tissues.
PCK2 and lung cancer TCGA database clinical parameters
Lung cancer patient clinical parameters were obtained from the TCGA database, and the link between PCK2 and these clinical indicators and prognosis was assessed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Creating nomograms and assessing their effectiveness
Nomograms were constructed using data from the multivariate analysis. The predictive ability of the nomogram was assessed with the use of a calibration curve.
Protein-gene interaction network diagram building using the GeneMania and STRING databases
The GeneMania database is used to generate assumptions about gene function, analyze gene lists, and prioritize genes for functional analysis. Given a list of query genes, GeneMania uses a large number of genomics and proteomics data to find genes with similar functions. Another use of GeneMania is for predicting gene function. Given a query gene, GeneMania will identify genes that may share functions with it based on their interactions. String database (https://string-db.org/). It is a database that searches for interactions between proteins. This includes both direct physical interactions between proteins and indirect functional correlations between proteins. Using GeneMania, we mapped out the genetic relationships between PCK2 genes and modified those in close proximity. The PCK2 protein-protein interaction (PPI) network was also generated using the STRING database for comparison.
Genome-wide analysis of PCK2 gene co-expression
Using data mining on the TCGA database, we determined which genes were positively or negatively co-expressed with PCK2.
PCK2 and immune cells: a link between the TIMER and TCGA databases
To visualize the association between PCK2 expression and different immune cells in lung adenocarcinoma, we built a stick figure through the TIMER database (https://cistrome.shinyapps.io/timer/).
Examining mutations in genes
Change frequency, mutation site information, a mutation type, and a 3D structure of all TCGA cancer candidate proteins were analysed in the cBioPortal database (including lung adenocarcinoma).
Sequencing of individual cells
CancerSEA is a specialized database for single-cell sequencing that may reveal a variety of functional states of cancer cells at the single-cell level. Single-cell sequencing data was used to study PCK2 expression and the performance of lung adenocarcinomas correlation. T-distributed Stochastic Neighbor Embedding (T-SNE) diagram shows the expression profile of PCK2 in single cells of TCGA samples.
Analyzing the UALCAN database to compare promoter methylation levels across lung cancer patients
To examine and contrast the promoter methylation levels of various lung adenocarcinoma patients, their information was entered into the UALCAN database for CPTAC analysis.
Gene set enrichment analysis (GSEA)
The biological role of PCK2 in lung cancer was investigated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. A potent bioinformatics method, GO analysis may be used to identify PCK2-related cellular components (CCs), molecular activities (MFs), and biological processes (BPs). The fundamental mechanism of PCK2 was investigated using GSEA (16).
Statistical analysis
Web resources were used to automatically compute the statistical analyses. Statistical significance was assumed at a P value or a log rank P value of <0.05.
Results
Downregulation of PCK2 expression in lung adenocarcinoma
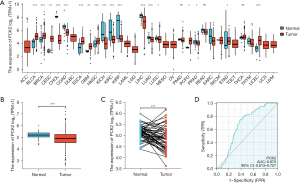
Pan-cancer analysis using the TCGA database revealed the expression of PCK2 was reduced in most tumors (Figure 1A). In the paired and unpaired tumor tissues of lung adenocarcinoma, paracancerous tissues had a greater PCK2 expression than cancerous ones (Figure 1B,1C), and the area under the curve (AUC) of the receiver operator characteristic (ROC) was 0.670. This result indicates PCK2 also has certain predictive research value (Figure 1D).
PCK2 and lung cancer prognosis in TCGA database
In the TCGA database, we discovered that patients with high expressions of PCK2 in their lung adenocarcinoma had superior overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) (Figure 2).

PCK2 expression in TCGA and lung cancer clinical data
Patients with reduced expression of PCK2 had a greater T stage, N stage, pathological stage, and pathological grade when compared to the clinical data of patients with lung adenocarcinoma, which is consistent with the expression of prior study findings (Figure 3).

Nomogram construction
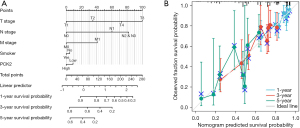
Using multivariate analytic findings, we built a nomogram from the TCGA database to predict survival rates for 1-, 3-, and 5-year in patients with lung adenocarcinoma. The C-index of the nomogram was 0.683 (0.658–0.707) (Figure 4A), and the calibration curve demonstrated it had a predictive value (Figure 4B).
Genes and proteins interacting with PCK2 were identified
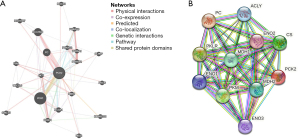
Using GeneMania, we formed a PCK2 gene-gene interaction network and modified neighboring genes (Figure 5A). To create the PPI network for PCK2, we queried the STRING database for protein-protein interactions (Figure 5B).
PCK2 co-expression gene screening
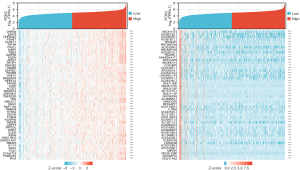
To find genes either positively or negatively linked with PCK2, we mined the TCGA database for information on lung cancer and compiled a list of the most 50 positively correlated (Figure 6A) and negatively correlated (Figure 6B) genes and displayed their expressions on a co-expression heat maps.
PCK2 and immunological cells: a TIMER and TCGA database correlation
Using the TIMER database, we created a diagram depicting the PCK2 expression and tumor purity correlation to different immune cells in lung adenocarcinoma. PCK2 expression was correlated positively to CD4+ T cells, B cells, macrophages, CD8+ T cells, dendritic cells, and neutrophils, which are six of the eight categories of invading immune cells (Figure 7A). The infiltration levels of PCK2 and aDC, Treg, iDC, TFH, DC, pDC, and Th2 cells were all positively correlated, but levels of infiltration for Tcm, T helper cells, CD8 T cells, and Th17 cells were negatively correlated. Accordingly, we performed a more in-depth analysis of the effect of PCK2 on the TME by analyzing the correlation between PCK2 and specific immune cells (Figure 7B). Additional research revealed a statistically significant positive link between PCK2 expression and PDCD1, a molecule associated with immunological checkpoints, although no such difference existed between PCK2 and CTLA4 or CD274 (Figure 7C). These results provide a foundation for further investigation and raise the possibility that PCK2 expression is linked to immune infiltration into lung adenocarcinoma tumors. They also suggest PCK2 plays a significant function in preventing the escape of immune by tumor cells within the lung adenocarcinoma tumor microenvironment.
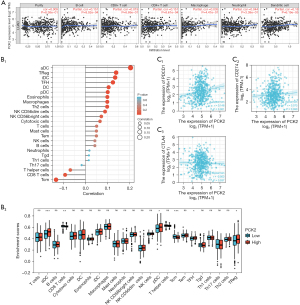
Mutations of PCK2 in lung adenocarcinoma
To further understand PCK2 gene mutation in cancer, we evaluated its various mutation statuses using the cBioPortal platform and TCGA data. In lung adenocarcinoma, the mutation rate of PCK2 was 0.53%, while the amplification rate was 1.77% and the deep deletion rate was 0.35%, according to a global study of cancer (Figure 8A). Figure 8B shows missenses and truncations were the main mutation types in PCK2, and Figure 8C shows the change at A477T/S in the 3D structure of the PCK2 protein.
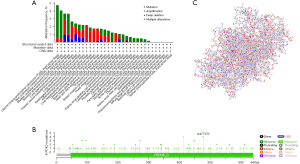
PCK2 promoter methylation in lung adenocarcinoma subtypes
Cancer and development may both be affected by promoter DNA methylation, which has been related to transcriptional suppression. In a stratified examination of lung adenocarcinoma patients, we found that PCK2 promoter methylation decreased with disease progression, and regardless of whether they had the TP53 mutation, patients fell into a wide range of age groups, N stages, and PCK2 promoter methylation patterns. Therefore, we can speculate that the expression of transcriptional PCK2 may be due to changes in promoter methylation (Figure 9).
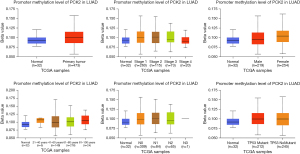
Single-cell analysis of PCK2 expression in lung adenocarcinoma
One of the most useful tools for assessing the possible roles of candidate molecules in a single cell is single-cell transcriptome sequencing. To start, we examined the correlation between PCK2 expression and phenotypes in a variety of tumor types using the CancerSEA database (including lung adenocarcinoma). Figure 10A shows that in lung cancer, PCK2 is negatively correlated with epithelial-mesenchymal transition (EMT) and hypoxia, and has other relationships with autophagy, cell cycle, differentiation, and inflammation (Figure 10B). By using T-SNE plots, we also displayed the PCK2 expression profile at the single-cell level in lung cancer (Figure 10C).
GO and KEGG enrichment analysis
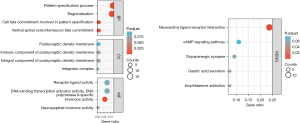
To evaluate the role of PCK2 in the development and progression of lung adenocarcinoma, we next performed a functional enrichment analysis. Gene ontology (GO) and KEGG enrichment analysis (Tables 1,2) showed genes co-expressed with PCK2 affect DNA-binding transcriptional activator activity, RNA polymerase II specificity, neuropeptide hormone activity, neuroactive ligand-receptor interaction, dopaminergic synapse, cAMP signaling pathway, and other features involved in the occurrence and development of lung adenocarcinoma (Figure 11).
Table 1
| Ontology | ID | Description | Gene ratio | Bg ratio | P value | Adjust P value | q value |
|---|---|---|---|---|---|---|---|
| BP | GO:0003002 | Regionalization | 16/144 | 351/18,670 | 1.35e-08 | 2.35e-05 | 2.15e-05 |
| BP | GO:0007389 | Pattern specification process | 17/144 | 446/18,670 | 6.36e-08 | 5.54e-05 | 5.07e-05 |
| BP | GO:0060579 | Ventral spinal cord interneuron fate commitment | 4/144 | 15/18,670 | 4.34e-06 | 0.002 | 0.002 |
| BP | GO:0060581 | Cell fate commitment involved in pattern specification | 4/144 | 15/18,670 | 4.34e-06 | 0.002 | 0.002 |
| CC | GO:0099061 | Integral component of postsynaptic density membrane | 4/153 | 50/19,717 | 6.08e-04 | 0.072 | 0.068 |
| CC | GO:0099146 | Intrinsic component of postsynaptic density membrane | 4/153 | 53/19,717 | 7.60e-04 | 0.072 | 0.068 |
| CC | GO:0032039 | Integrator complex | 3/153 | 28/19,717 | 0.001 | 0.082 | 0.078 |
| CC | GO:0098839 | Postsynaptic density membrane | 4/153 | 74/19,717 | 0.003 | 0.096 | 0.091 |
| MF | GO:0005179 | Hormone activity | 9/137 | 122/17,697 | 4.46e-07 | 1.15e-04 | 1.04e-04 |
| MF | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 11/137 | 439/17,697 | 6.21e-04 | 0.080 | 0.072 |
| MF | GO:0005184 | Neuropeptide hormone activity | 3/137 | 28/17,697 | 0.001 | 0.080 | 0.072 |
| MF | GO:0048018 | Receptor ligand activity | 11/137 | 482/17,697 | 0.001 | 0.080 | 0.072 |
GO, Gene Ontology; PCK2, phosphoenolpyruvate carboxykinase 2; BP, biological process; CC, cellular component; MF, molecular activity.
Table 2
| Ontology | ID | Description | Gene ratio | Bg ratio | P value | Adjust P value | q value |
|---|---|---|---|---|---|---|---|
| KEGG | hsa04080 | Neuroactive ligand-receptor interaction | 13/55 | 341/8,076 | 3.14e-07 | 3.92e-05 | 3.37e-05 |
| KEGG | hsa05031 | Amphetamine addiction | 4/55 | 69/8,076 | 0.001 | 0.061 | 0.052 |
| KEGG | hsa04971 | Gastric acid secretion | 4/55 | 76/8,076 | 0.002 | 0.061 | 0.052 |
| KEGG | hsa04728 | Dopaminergic synapse | 5/55 | 132/8,076 | 0.002 | 0.061 | 0.052 |
| KEGG | hsa04024 | cAMP signaling pathway | 6/55 | 216/8,076 | 0.003 | 0.075 | 0.064 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; PCK2, phosphoenolpyruvate carboxykinase 2.
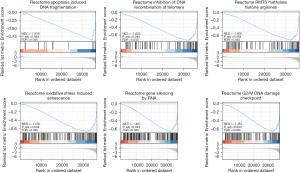
GSEA-based pathway prediction in signal transduction
To analyze the functionality of genes, we used Metascape to perform KEGG functional analysis online. This revealed PCK2 may have an impact on lung cancer development by taking part in the reaction to oxidative stress-induced senescence, gene silencing, cell cycle, reactive protein methylation of histone arginine, suppression of telomere responses, and DNA damage caused by apoptosis. This suggests that in lung adenocarcinoma, the prognosis might vary according to biological factors.
In light of the aforementioned prediction findings and the molecular properties of PCK2, we hypothesize it is a therapeutic target that causes senescence through the oxidative stress response and prevents the immunological escape of tumor cells. The experimental foundation established serves as a standard for further research (Figure 12).
Discussion
Lung cancer diagnoses numbered 228,000 in the United States in 2019 with around 160,000 deaths (17), demonstrating the disease has a high incidence, high fatality rate, and a poor prognosis. Lung adenocarcinoma is the most prevalent form of NSCLC (18,19), and determining its mechanism of incidence, invasion, and metastasis. Nowadays, with many technical breakthroughs in disciplines such as bioinformatics, molecular biology, and immunology, molecular targeted therapy, immunosuppressant therapy, and other methods have been widely used in clinical practice. However, there has been no notable improvement in the survival rate for those with lung adenocarcinoma (20,21). Therefore, it is crucial to better understand the causes of the disease to identify useful biomarkers and create effective novel treatments.
Abnormal cell metabolism is a key feature of tumor development, and abnormal glucose metabolism is a basic cellular function. While targeting aerobic glycolysis in tumor cells is a promising therapeutic strategy attracting much research interest, there are fewer studies on gluconeogenesis (22). Gluconeogenesis may be thought of as the inverse of glycolysis, with its three stages mirroring the three phases of the latter. To begin, pyruvate is converted into phosphoenolpyruvate (PEP). Physiologically, the pyruvate kinase-catalyzed process in glycolysis is irreversible because its activation energy, or ΔG, is −7.5 kcal. Therefore, gluconeogenesis can only be stimulated by the presence of two high-energy links (23).
New sequencing and omics technologies have opened new avenues for researchers to learn the causes of lung adenocarcinoma and find potential treatments for the disease (24). Using bioinformatics, we hypothesized PCK2 may play a part in the etiology of lung cancer in this investigation. Patients with high PCK2 expression in the TCGA database had higher DSS, OS, and PFS when compared to those with a low expression. Using the database’s multivariate analysis findings, we built a prediction model with a certain degree of accuracy that can potentially predict the 1-, 3-, and 5-year survival probability of lung adenocarcinoma patients. The expression of PCK2 was shown to be positively connected with six different kinds of invading immune cells in the TIMER database, including CD4+ T cells, B cells, macrophages, neutrophils, CD8+ T cells, and dendritic cells. The PDCD1 expression was positively connected with PCK2, and PCK2 in turn was positively correlated with the infiltration levels of aDC, Treg, iDC, TFH, DC, pDC, and Th2 cells. Although there is currently no clear literature suggesting experimental confirmation of PCK2 and immune infiltration. But it may indicate that LUAD in tumor immune microenvironment is regulated by PCK2. It has been suggested that the tumor microenvironment of lung cancer consists of tumor associated macrophages (TAM), a small number of invasive dendritic cells and natural killer cell. Previous studies have thoroughly studied the role of tumor associated T cells in the development of lung cancer. The factors influencing the prognosis of LUAD are the activation of CD4+ Th1 cells and activated CD8+ T cells, which enhance the immune response of the body (25,26). These findings suggest PCK2 may play a critical role in the tumor microenvironment of lung adenocarcinoma by preventing tumor cells from evading the immune system. We then examined the role of PCK2 mutations in lung adenocarcinoma and discovered missense and truncation mutations account for most PCK2 changes in this cancer. In addition to MNNG HOS transforming gene (c-MET) amplification, C797S mutation, and ERK pathway inhibition, previous research suggests epidermal growth factor receptor membrane/cytoplasmic/nuclear translocation may be an important cause of drug resistance in lung adenocarcinoma (27). Additionally, KRAS-G12D mutation can drive immunosuppression and enhance ICI resistance in NSCLC (28). We observed the methylation level of the PCK2 promoter reduced as the pathological stage of lung adenocarcinoma patients progressed, and upon the expression pattern at the single-cell level, PCK2 was seen to be negatively linked with EMT and hypoxia.
To summarise, we conducted a predictive analysis of related mechanism pathways, finding PCK2 may play an important role in the response to oxidative stress-induced senescence, gene silencing, the cell cycle, reactive protein methylation of histone arginine, inhibition of telomere response, and apoptosis-induced DNA damage, all of which affect biological events in lung adenocarcinoma. Cancer researchers have identified cell cycle arrest as a major feature of cellular senescence that must be overcome. The prevention of EMT, tumor suppression, or tumor advancement are all outcomes of cellular senescence, which may be triggered by a wide range of stimuli such as telomere shortening, oncogenic activation, and treatment. Restoring epigenetically silenced PCK2 inhibits renal cell carcinoma by promoting endoplasmic reticulum stress progression and increased sensitivity to sunitinib (29). Liu et al. (30) discovered that metabolic reprogramming of PCK1 and PCK2 promoted TCA catalysis, oxidative stress, and apoptosis, suggesting low expression levels of PCK2 may contribute to better prognosis by triggering the senescent
Conclusions
Accumulating evidence suggests PCK2 plays an important role in lung adenocarcinoma and has potential as a biomarker of disease progression through many mechanisms. Prediction findings and PCK2 molecular features suggest PCK2 improves the prognosis of lung adenocarcinoma patients by inducing senescence through oxidative stress and inhibiting the immune escape of tumor cells. Further cytological investigations are required for appropriate verification and research, but the finding suggests a potential target for anticancer therapies for the effective treatment of lung adenocarcinoma.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-542/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-542/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-542/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ott S, Geiser T. Epidemiology of lung tumors. Ther Umsch 2012;69:381-8. [Crossref] [PubMed]
- Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288-300. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Gelsomino F, Lamberti G, Parisi C, et al. The evolving landscape of immunotherapy in small-cell lung cancer: A focus on predictive biomarkers. Cancer Treat Rev 2019;79:101887. [Crossref] [PubMed]
- Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 2018;9:117. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Wang Z, Dong C. Gluconeogenesis in Cancer: Function and Regulation of PEPCK, FBPase, and G6Pase. Trends Cancer 2019;5:30-45. [Crossref] [PubMed]
- Huang J, Tian F, Song Y, et al. A feedback circuit comprising EHD1 and 14-3-3zeta sustains beta-catenin/c-Myc-mediated aerobic glycolysis and proliferation in non-small cell lung cancer. Cancer Lett 2021;520:12-25. [Crossref] [PubMed]
- Zhang W, Zhang SL, Hu X, et al. Targeting Tumor Metabolism for Cancer Treatment: Is Pyruvate Dehydrogenase Kinases (PDKs) a Viable Anticancer Target? Int J Biol Sci 2015;11:1390-400. [Crossref] [PubMed]
- Smolle E, Leko P, Stacher-Priehse E, et al. Distribution and prognostic significance of gluconeogenesis and glycolysis in lung cancer. Mol Oncol 2020;14:2853-67. [Crossref] [PubMed]
- Benny S, Mishra R, Manojkumar MK, et al. From Warburg effect to Reverse Warburg effect; the new horizons of anti-cancer therapy. Med Hypotheses 2020;144:110216. [Crossref] [PubMed]
- Stark R, Kibbey RG. The mitochondrial isoform of phosphoenolpyruvate carboxykinase (PEPCK-M) and glucose homeostasis: has it been overlooked? Biochim Biophys Acta 2014;1840:1313-30. [Crossref] [PubMed]
- Bluemel G, Planque M, Madreiter-Sokolowski CT, et al. PCK2 opposes mitochondrial respiration and maintains the redox balance in starved lung cancer cells. Free Radic Biol Med 2021;176:34-45. [Crossref] [PubMed]
- Frodyma DE, Troia TC, Rao C, et al. PGC-1beta and ERRalpha Promote Glutamine Metabolism and Colorectal Cancer Survival via Transcriptional Upregulation of PCK2. Cancers (Basel) 2022;14:4879. [Crossref] [PubMed]
- Luo S, Li Y, Ma R, et al. Downregulation of PCK2 remodels tricarboxylic acid cycle in tumor-repopulating cells of melanoma. Oncogene 2017;36:3609-17. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Xu X, Zhu H, Yang M, et al. Knockdown of TOR signaling pathway regulator suppresses cell migration and invasion in non-small cell lung cancer via the regulation of epithelial-to-mesenchymal transition. Exp Ther Med 2020;19:1925-32. [PubMed]
- Cheng WC, Chang CY, Lo CC, et al. Identification of theranostic factors for patients developing metastasis after surgery for early-stage lung adenocarcinoma. Theranostics 2021;11:3661-75. [Crossref] [PubMed]
- Zhang S, Chen Z, Shi P, et al. Downregulation of death receptor 4 is tightly associated with positive response of EGFR mutant lung cancer to EGFR-targeted therapy and improved prognosis. Theranostics 2021;11:3964-80. [Crossref] [PubMed]
- Wu Q, Yu L, Lin X, et al. Combination of Serum miRNAs with Serum Exosomal miRNAs in Early Diagnosis for Non-Small-Cell Lung Cancer. Cancer Manag Res 2020;12:485-95. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Liu J, Li Y, Lu Z, et al. Deceleration of glycometabolism impedes IgG-producing B-cell-mediated tumor elimination by targeting SATB1. Immunology 2019;156:56-68. [Crossref] [PubMed]
- Hatting M, Tavares CDJ, Sharabi K, et al. Insulin regulation of gluconeogenesis. Ann N Y Acad Sci 2018;1411:21-35. [Crossref] [PubMed]
- Dhanasekaran R, Nault JC, Roberts LR, et al. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology 2019;156:492-509. [Crossref] [PubMed]
- Bremnes RM, Busund LT, Kilvær TL, et al. The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:789-800. [Crossref] [PubMed]
- Schalper KA, Brown J, Carvajal-Hausdorf D, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 2015;107:dju435. [Crossref] [PubMed]
- Rong X, Liang Y, Han Q, et al. Molecular Mechanisms of Tyrosine Kinase Inhibitor Resistance Induced by Membranous/Cytoplasmic/Nuclear Translocation of Epidermal Growth Factor Receptor. J Thorac Oncol 2019;14:1766-83. [Crossref] [PubMed]
- Liu C, Zheng S, Wang Z, et al. KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun (Lond) 2022;42:828-47. [Crossref] [PubMed]
- Xiong Z, Yuan C, Shi J, et al. Restoring the epigenetically silenced PCK2 suppresses renal cell carcinoma progression and increases sensitivity to sunitinib by promoting endoplasmic reticulum stress. Theranostics 2020;10:11444-61. [Crossref] [PubMed]
- Liu MX, Jin L, Sun SJ, et al. Metabolic reprogramming by PCK1 promotes TCA cataplerosis, oxidative stress and apoptosis in liver cancer cells and suppresses hepatocellular carcinoma. Oncogene 2018;37:1637-53. [Crossref] [PubMed]
(English Language Editor: B. Draper)


