Assessment of epidermal growth factor receptor mutation/copy number and K-ras mutation in esophageal cancer
Introduction
Esophageal cancer is the eighth most common cancer worldwide, with an estimated 482,300 new esophageal cancer cases and 406,800 deaths occurred in 2008 worldwide (1,2). In China, esophageal cancer is the fourth most common cause of cancer-related death, and the mortality is 10.97/100,000 (7.31%) in urban areas and 17.34/100,000 (13.48%) in country areas, respectively (3). Although great improvement has been made in multi-modality approaches, the 5-year survival rate for esophageal cancer patients is only 17% (4).
Targeted therapy, designed rationally to inhibit appropriate signaling pathways in human solid tumor, may provide effective, highly selective, and well-tolerated anticancer treatments, the patients with epidermal growth factor receptor (EGFR) exon 19 or 21 mutations are known to receive more benefits from gefitinib therapy in clinical practice (5). In addition, increased gene copy number is another mechanism of oncogene activation. Dahabreh et al. (6) and Hirsch et al. (7) have reported that the increased EGFR gene copy number is also a predictive biomarker for response to TKIs in non-small cell lung cancer (NSCLC), and EGFR gene amplification is in association with tumor progression of EGFR-mutated NSCLC. Both mutation and gene amplification are probably important in determining TKIs sensitivity. On the other hand, it was later evidenced that a proportion of patients with EGFR mutation did not respond to therapy with EGFR inhibitors, and the presence of activating K-ras mutation has been identified as a potent predictor of resistance to EGFR-inhibitors. The TKIs should therefore be applied only in tumors with a wild-type status of the K-ras gene (8).
Given the significant implication of EGFR in multiple steps of the progression toward solid tumors, it is not surprising that EGFR and its associated pathways may become an active area of investigation for targeted therapeutics in esophageal cancer. Deep elucidation for status of EGFR, K-ras mutations, as well as EGFR gene copy number is an indispensable step for exploring esophageal cancer targeted therapy with TKIs. Previous studies have implicated EGFR over-expression, tested by immunohistochemistry (IHC) method in esophageal adenocarcinoma (EADC), being associated with higher pathologic stage and poor prognosis (9,10). However, limited studies focused on the EGFR gene copy number and the identification of activating mutations, especially in esophageal squamous cell carcinoma (ESCC). Here, we detected the EGFR and K-ras mutation, as well as the EGFR gene copy number status in esophageal cancer, and investigated the relationship between EGFR gene copy number status and clinicopathologic features or patients’ survival.
Methods
Patients and tumor samples
This study enrolled 66 consecutive patients with clinically localized ESCC or adenocarcinoma who had undergone three field esophagectomy from 2009 to 2011. All patients signed an informed consent for the use of their fresh tissues, and the study was approved by the institutional review board of the Cancer Center of Sun Yat-Sen University. Fresh tissues were procured from surgical resection specimens collected by department of pathology in the Cancer Center of Sun Yat-Sen University. The condition of the patients was assessed according to the system for staging primary tumor, node, metastases (TNM) described in the American Joint Committee on Cancer Staging Manual (11). The patients who had received preoperative chemotherapy or preoperative chemo-radiotherapy were excluded in this study.
Real-time PCR analysis of EGFR and K-RAS mutations
Genomic DNA was extracted from the fresh tumor tissue with a QIAamp DNA mini kit (GP Medical Technologies Ltd, Beijing, China) according to the manufacturer’s instructions. We used an EGFR Mutation Detection Kit (GP Medical Technologies Ltd) to detect a deletion in exon 19 (delE746A750) and mutation in exon 21 (L858R) by real-time polymerase chain reaction (RT-PCR) (12). The reaction conditions were as follows: initial activation of DNA polymerase at 50 °C for 2 min, denaturation at 95 °C for 10 min, 40 cycles of amplification at 95 °C for 15 s and at 62 °C for 60 s. We also used K-ras mutation Detection Kit (GP Medical Technologies Ltd) to detect codons 12/13 mutation by RT-PCR (13). PCR conditions for K-ras mutation were as follows: 1 cycle at 95 °C for 9 min, 45 cycles at 94 °C for 1 min, at 55 °C for 1 min, and at 72 °C for 1 min, followed by 1 cycle at 72 °C for 5 min.
Fluorescent in situ hybridization (FISH) analysis of EGFR gene copy number
FISH assays were performed using the EGFR FITC Red/CEP 7 Rhodamine Green probe (GP Medical Technologies Ltd) according to the manufacturer’s instructions. FISH analyses were defined according to the previously published criteria by Cappuzzo and his colleagues (14). Six FISH strata were classified according to the frequency of tumor cells with specific number of copies of the gene and chromosome 7 centromere: (I) disomy (≤2 copies of per cell in >90% of cells); (II) low trisomy (≤3 copies in 10–40%of cells); (III) high trisomy (≤3 copies in >40% of cells); (IV) low polysomy (≥4 copies in 10–40% of cells); (V) high polysomy (≥4 copies in >40% of cells); (VI) gene amplification (tight gene clusters and a ratio of gene to chromosome of ≥2, or ≥10 copies in ≥10% of cells). Gene amplification and high polysomy were defined as high EGFR copy number status or FISH-positive and all else were defined as low EGFR copy number status or FISH-negative.
Statistical analysis
Statistical analysis was performed using SPSS software (version 13 for Windows; SPSS, Chicago, IL, USA). The χ2 test or Fisher’s exact test were employed to examine the correlation between the status of EGFR gene copy number and the clinicopathologic characteristics. For univariate analysis, survival curves were obtained with the Kaplan-Meier method, and the log-rank test was employed to compare the difference within groups. Multivariate survival analysis was performed with the cox proportional hazards regression model. The corresponding hazard ratio (HR) and 95% CI were taken from Cox regression models. We used a 2-sided significance level of P<0.05 for all statistical analyses.
Results
EGFR mutation
The clinicopathologic characteristics of the study population are summarized in Table 1. All the tumors were completely resected (R0 category), and the median survival time was 36 months. We detected EGFR mutations in all the patients. The result revealed that only one patient had EGFR mutation with exon 19 in-frame deletion. He was diagnosed as mid-esophageal carcinomas, with pathological stage IIIC disease. The histopathological examination showed a high-differentiated ESCC with tumor invasion of esophageal adventitia and 7 of 27 lymph nodes metastasis.
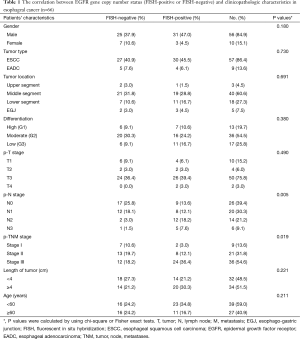
Full table
K-ras mutation
The K-ras mutation was also identified in the all the patients, only one case was found to be activating mutant, representing a glycine to aspartic acid substitution at codon 12. The patient who had this mutation was treated by esophagectomy with three field lymphadenectomy. The pathological diagnosis was low-differentiated EADC; and the depth of the tumor was infiltrating into the submucosa. Total 16 lymph nodes were dissected, and neither lymph node positive nor distant metastasis was verified.
EGFR gene copy number status
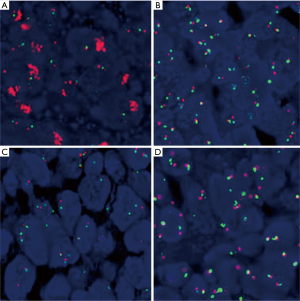
EGFR gene copy number status was analyzed using FISH methods in 66 specimens. Four major FISH patterns (normal disomy, low trisomy, high polysomy, and gene amplification) were detected out in the study and illustrated in Figure 1. There were 9 tumor samples with gene amplification (13.64%), 25 with high polysomy (37.9%), 16 with low polysomy (24.2%), and 16 with normal disomy (24.2%). Therefore, High EGFR gene copy number status (FISH-positive) was presented in 34 patient specimens (51.5%) and 32 specimens (48.5%) were low EGFR gene copy number status (FISH- negative).
The correlation between EGFR gene copy number status and patients’ characteristics, such as age, sex, tumor type, tumor location, tumor differentiation, tumor invasion (p-T stage), pathological lymph node stage (p-N stage), pathologic-TNM stage (p-TNM stage) and the length of tumor were investigated (Table 1). The results showed that high EGFR gene copy number status (FISH-positive) was significantly associated with advanced p-TNM stage (P=0.019) and more number of lymph node metastasis (P=0.005) in esophageal cancer (Table 1). Because of the majority of the patients are ESCC patients in the study, detailed analysis of ESCC patients was also performed. The results also showed that EGFR gene copy number status was also correlated with p-TNM stage (P=0.007) and lymph node metastasis (P=0.008) in ESCC patients (Table 2).
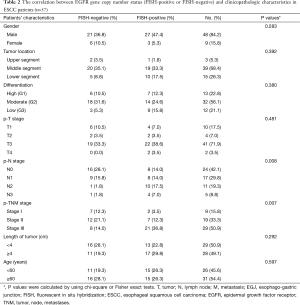
Full table
Survival analysis
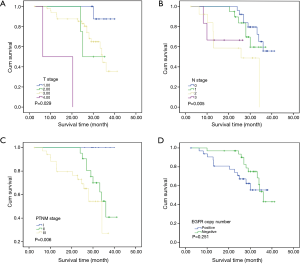
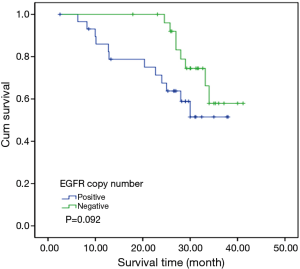
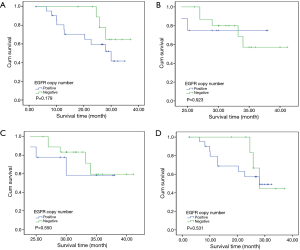
To investigate the relationship between clinical outcome of esophageal cancer patients and the clinicopathologic parameters, including the EGFR gene copy number status, survival curve was drawn by using Kaplan-Meier method, and regression analysis was performed by using the Cox’s proportional hazards model. The results showed that the pathological T stage (P=0.029), p-N stage (P=0.005) and p-TNM stage (P=0.006) were significant predictors of a poor overall survival (OS) (Table 3 and Figure 2A-C); At the same time, the results showed that the high EGFR gene copy number status was not a significant predictor of a poor outcome among esophageal cancer patients in the present study (P=0.251), However, the survival curve on EGFR gene copy number status showed a trend that esophageal cancer patients with low EGFR gene copy number may have longer survival than those with higher EGFR gene copy number (Figure 2D). Due to the majority of enrolled patients were ESCC patients, we then analyzed the effect of EGFR gene copy number status on ESCC patients’ survival, separately. The new survival curve showed a trend that ESCC patients with low EGFR gene copy number have a longer survival than those with higher EGFR gene copy number (P=0.092, Figure 3). These results indicated that the high EGFR gene copy number may have a deleterious effect on prognosis, although it makes no statistical significance. In addition, due to high EGFR gene copy number status was significantly associated with advanced p-TNM stage and more number of lymph node metastasis in ESCC patients, and it is a matter of course that advanced patients have a shorter OS. So we did the subgroup analyses in ESCC, which excluded the effects of p-TNM stage or p-N stage on OS, to evaluate the EGFR gene copy number’s effect on ESCC patients’ survival. Among p-N1-3 ESCC patients, or N0 ESCC patients, or p-TNM stage ESCC I/II patients, or p-TNM stage III ESCC patients, high EGFR gene copy number patients seems always showed a shorter OS compared to the patients with low EGFR gene copy number according the survival curve trends (Figure 4), although they made no statistical significance.
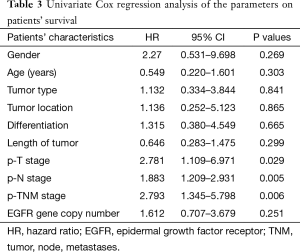
Full table
Discussion
EGFR gene amplification, mutation and over-expression are frequent in malignancy. EGFR protein expression was detected in 40–80% of NSCLCs (15,16), and 88.4% of over-expression of EGFR has been described in head and neck squamous cell carcinomas (HNSCCs) (17). In esophageal cancer, a wide range of EGFR expression has been reported: 32–85% of EADC and 12–71% of ESCC (18).
The tumor areas containing gene amplification was highly correlated with the areas of protein over-expression by IHC. Several studies reported that EGFR protein expression, EGFR copy number and EGFR mutations were closely related to each other in NSCLCs (19,20). Although EGFR copy number is less correlated with TKIs responsiveness when compared with EGFR mutations, it still could be a good alternative molecular predictive marker for TKIs responsiveness (21). There was a strong correlation between EGFR protein expression assessed by IHC and gene copy number assessed by FISH. Remarkably, Hanawa et al. also demonstrated a correlation between increasing levels of EGFR protein expression by IHC and increased gene copy number, and regions with EGFR amplification in esophageal cancer coincided with those exhibiting 2+ or 3+ degrees of immunoreactivity by IHC. The result suggests that the addictive effect of gene amplification or chromosome polysomy indicates a relevant mechanism underlying protein expression (22).
In the present study, high EGFR gene copy number was found in 51.5% of the study population, and was significantly associated with tumor stage (p-TNM) and locoregional lymph node metastasis. Our results are consistent with several previous studies that showed association between EGFR gene copy number and histopathological factors (22,23). Wilkinson found that the EGFR expression was correlated with histologic grade in resected EADC (18). Gibault et al. and Wang et al. respectively reported that EGFR was associated with reduced OS in locally ESCC (24,25). But we found no correlation between EGFR gene copy number status and age, gender, tumor location, histological grade. The EGFR gene copy number may play an important role in the esophageal development. The results indicate that evaluation of gene changes of EGFR in esophageal cancer is essential for individual prediction of the disease course and development of new approaches to the treatment of these tumors, including target therapy aimed at these tyrosine kinase receptors.
Activating EGFR mutations include in-frame deletions and amino acid substitutions in exons 18, 19, and 21. Mutations in the tyrosine kinase domain of EGFR were reported in the majority of tumors with dramatic responses to EGFR-targeted therapies, and an activating mutation of the EGFR tyrosine kinase domains was shown to be the most important factor predictive of the responses to TKI (26-28). In our study, there is only one patient detected with EGFR mutation, which indicates that the EGFR gene mutation is rare in esophageal cancer. Our findings are consistent with the previous study. Pühringer-Oppermann and his colleagues detected exons 19 and 21 coding of the EGFR gene in 105 samples of EADC by denaturing high-pressure liquid chromatography, resulting only one sample indicating a mutation (29). Hanawa et al. detected exons 19 and 21 of EGFR gene by direct sequencing, revealing no mutations in 40 tumor samples (22). Yong Cui identified exon 19 and 21 mutations of EGFR gene in Chinese patients with ESCC. No EGFR gene mutation was observed in the 127 tumor samples by direct sequencing method, but K-ras mutation was detected in 2 out of 127 (1.6%) patients (30).
The presence of K-ras mutation seems to be correlated with primary resistance to TKIs. Activating K-ras gene point mutation has been detected in many types of human tumors. Such as oncogenic forms of the K-ras gene were prevalent in pancreatic carcinomas (>80% of cases) (31), colon carcinomas (about 50% of cases) (32), and appeared in 10–30% of lung carcinoma cases (33). K-ras mutation has emerged as an important predictive marker of resistance to TKI treatment. So it is necessary to identify the mutation status of K-ras to select patients who may profit from target therapy. In our study, there was only one patient. The results were consistent with other studies in esophageal cancer, implying that the K-ras mutation in esophageal cancer is rare.
To our knowledge, there have been relatively few researches investigating the relationships between clinical features and EFGR gene copy number, and the present study is the first to focus on the relationships between clinical outcome and EGFR gene copy number status. Interestingly, when we analyzed our results, patients with a high EGFR gene copy number were found to have a shorter OS than those with low EGFR gene copy number, indicating that a high EGFR gene copy number is likely to have a deleterious effect on prognosis of esophageal cancer patients or ESCC patients, although the p value had no statistical significance. In addition, high EGFR gene copy numbers are more frequently encountered in advanced stage esophageal cancer patients, or ESCC patients. Our results indicate that further investigations are required to determine whether high EGFR gene copy number could be viewed as an independent poor prognostic factor in esophageal cancer or ESCC.
Conclusions
Our results indicated that EGFR or K-ras mutation was rare in esophageal cancer, but high EGFR gene copy number is frequent, and correlated with advanced pathologic stage and more number of the metastatic regional lymph nodes, especially in ESCC. In addition, high EGFR gene copy number is likely to have a deleterious effect on prognosis of esophageal cancer patients or ESCC patients, although no statistical significance was reached in the study.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the institutional review board of the Cancer Center of Sun Yat-Sen University and written informed consent was obtained from all patients.
References
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Zhao P, Dai M, Chen W, et al. Cancer trends in China. Jpn J Clin Oncol 2010;40:281-5. [Crossref] [PubMed]
- Shahbaz Sarwar CM, Luketich JD, Landreneau RJ, et al. Esophageal cancer: an update. Int J Surg 2010;8:417-22. [Crossref] [PubMed]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [Crossref] [PubMed]
- Dahabreh IJ, Linardou H, Siannis F, et al. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2010;16:291-303. [Crossref] [PubMed]
- Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol 2008;26:3351-7. [Crossref] [PubMed]
- Raponi M, Winkler H, Dracopoli NC. KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol 2008;8:413-8. [Crossref] [PubMed]
- Wang KL, Wu TT, Choi IS, et al. Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas: association with poor outcome. Cancer 2007;109:658-67. [Crossref] [PubMed]
- Li JC, Zhao YH, Wang XY, et al. Clinical significance of the expression of EGFR signaling pathway-related proteins in esophageal squamous cell carcinoma. Tumour Biol 2014;35:651-7. [Crossref] [PubMed]
- Yam PC, Tong D, Law S. Comparisons of sixth and seventh edition of the American Joint Cancer Committee staging systems for esophageal cancer. Ann Surg Oncol 2014;21:583-8.
- Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 2006;12:3915-21. [Crossref] [PubMed]
- Aoki Y, Hosaka S, Tachibana N, et al. Reassessment of K-ras mutations at codon 12 by direct PCR and sequencing from tissue microdissection in human pancreatic adenocarcinomas. Pancreas 2000;21:152-7. [Crossref] [PubMed]
- Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 2005;97:643-55. [Crossref] [PubMed]
- Herbst RS, Shin DM. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors: a new paradigm for cancer therapy. Cancer 2002;94:1593-611. [Crossref] [PubMed]
- Sridhar SS, Seymour L, Shepherd FA. Inhibitors of epidermal-growth-factor receptors: a review of clinical research with a focus on non-small-cell lung cancer. Lancet Oncol 2003;4:397-406. [Crossref] [PubMed]
- Goldstein NS, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: implications for a standardized scoring system. Cancer 2001;92:1331-46. [Crossref] [PubMed]
- Wilkinson NW, Black JD, Roukhadze E, et al. Epidermal growth factor receptor expression correlates with histologic grade in resected esophageal adenocarcinoma. J Gastrointest Surg 2004;8:448-53. [Crossref] [PubMed]
- Liang Z, Zhang J, Zeng X, et al. Relationship between EGFR expression, copy number and mutation in lung adenocarcinomas. BMC Cancer 2010;10:376. [Crossref] [PubMed]
- Zhang LJ, Cai L, Li Z, et al. Relationship between epidermal growth factor receptor gene mutation and copy number in Chinese patients with non-small cell lung cancer. Chin J Cancer 2012;31:491-9. [Crossref] [PubMed]
- Chang JW, Liu HP, Hsieh MH, et al. Increased epidermal growth factor receptor (EGFR) gene copy number is strongly associated with EGFR mutations and adenocarcinoma in non-small cell lung cancers: a chromogenic in situ hybridization study of 182 patients. Lung Cancer 2008;61:328-39. [Crossref] [PubMed]
- Hanawa M, Suzuki S, Dobashi Y, et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer 2006;118:1173-80. [Crossref] [PubMed]
- Kitagawa Y, Ueda M, Ando N, et al. Further evidence for prognostic significance of epidermal growth factor receptor gene amplification in patients with esophageal squamous cell carcinoma. Clin Cancer Res 1996;2:909-14. [PubMed]
- Gibault L, Metges JP, Conan-Charlet V, et al. Diffuse EGFR staining is associated with reduced overall survival in locally advanced oesophageal squamous cell cancer. Br J Cancer 2005;93:107-15. [Crossref] [PubMed]
- Wang Q, Zhu H, Xiao Z, et al. Expression of epidermal growth factor receptor is an independent prognostic factor for esophageal squamous cell carcinoma. World J Surg Oncol 2013;11:278. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol 2005;23:6829-37. [Crossref] [PubMed]
- Pühringer-Oppermann FA, Stein HJ, Sarbia M. Lack of EGFR gene mutations in exons 19 and 21 in esophageal (Barrett's) adenocarcinomas. Dis Esophagus 2007;20:9-11. [Crossref] [PubMed]
- Cui Y, Chang D, Liu M, et al. Identification of exon 19 and 21 mutations of EGFR gene in Chinese patients with esophageal squamous cell carcinoma. World J Surg Oncol 2013;11:266. [Crossref] [PubMed]
- Li D, Firozi PF, Zhang W, et al. DNA adducts, genetic polymorphisms, and K-ras mutation in human pancreatic cancer. Mutat Res 2002;513:37-48. [Crossref] [PubMed]
- Bazan V, Agnese V, Corsale S, et al. Specific TP53 and/or Ki-ras mutations as independent predictors of clinical outcome in sporadic colorectal adenocarcinomas: results of a 5-year Gruppo Oncologico dell'Italia Meridionale (GOIM) prospective study. Ann Oncol 2005;16 Suppl 4:iv50-55. [Crossref] [PubMed]
- Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer 2001;92:1525-30. [Crossref] [PubMed]


