Positive expression of miR-361-5p indicates better prognosis for breast cancer patients
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer among women in China and worldwide (1,2). It is considered to be a heterogeneous disease comprising four subtypes (luminal A, luminal B, HER2+, and basal-like) which has different molecular characteristics, behaviors, and response to treatment (3). Individual molecular markers were proved to be associated with clinical diagnosis, prognosis and predictions in treatment responses in BC. And numerous biomarkers have been validated and evaluated for clinical application (4-7), such as PAM50 assay (based on the NanoString technology), MammaPrint test (based on the microarray technology), Oncotype DX test (based on the qRT-PCR technology).
MicroRNAs (miRNAs) are none-coding small RNAs which are 18–24 nucleotides in length, working at the post-transcriptional level of the gene expression regulation (8). miRNAs are important regulators of diverse critical cellular processes including cell cycle regulation, cell growth, apoptosis, cell differentiation and stress response (9). Increasing evidence indicated that aberrant miRNA expression was associated with tumor invasion, distal metastasis and resistance to chemotherapy. Thereby, miRNA expression signatures are considered to be promising predictive and prognostic biomarkers for BC (10,11). Increasing studies independently reported onco-miR such as miR-21, miR-210 were over-expressed in BC and the expression levels correlated with tumor aggressiveness and poor prognosis (12). In contract, high expression of miRNAs as tumor suppressors such as miR-30e, let-7b were associated with good prognosis (13,14).
MiR-361-5p, which is located on chromosome X, has been reported to play a critical role in several human tumors. It has been demonstrated to be down-regulated in colorectal carcinoma and gastric cancer, suggesting that miR-361-5p functioned as a tumor-suppressive miRNA (15). Decreased miR-361-5p expression has also been observed in castration-resistant prostate cancer as well (16). However, a contradictory report identified that miR-361-5p acted as an oncogene in cervical cancer progression by enhancing cell proliferation and invasion (17). These conflicting research results of miR-361-5p in different tumors necessitate the validation of the precise role of this miRNA in specific tumors. To our knowledge, no previous research has focused on the expression of miR-361-5p and its exact role in BC. Therefore, we aimed to examine the expression level of miR-361-5p in BC and investigate the specific function and potential prognostic value.
Methods
Study population
In this study, data and specimens were collected from 375 female patients who were diagnosed with stage I to III BC at the Department of Breast Surgery in Fudan University Shanghai Cancer Center (FUSCC, Shanghai, China) between August 2001 and March 2005. All patients were histologically confirmed as invasive ductal breast carcinoma. The follow-up time was measured from the date of surgery until patient’s death or last follow-up. Patients were regularly followed-up and the median follow-up time was 96.57 months. The study was approved by the Ethics Committee of FUSCC, and a written informed consent was signed by each patient.
Tissue microarrays (TMAs)
The TMAs were constructed from formalin-fixed and paraffin-embedded (FFPE) samples from above BC patients before their cancer treatment by the Department of Pathology in FUSCC. Representative tumor regions were marked and tissue cylinders were then punched from these regions and transferred to recipient array blocks by TMA. The TMAs were constructed in duplicate so as to compare the staining in different areas of the same tumor.
In situ hybridization (ISH)
ISH was performed on the TMAs using the digoxigenin (DIG)-labeled miRCURY LNATM microRNA detection probes for miR-361-5p (Exiqon, Vedbeak, Denmark) and the Enhanced Sensitive ISH Detection Kit I (POD) (Boster Company, Wuhan, China). Briefly, the ISH was performed as follows (18): The TMA slides were incubated at 68 °C for 4 h. They were then deparaffinized in xylene, hydrated in graded ethanol series and washed by phosphate buffered saline (PBS). Endogenous peroxidase activity was quenched with 3% H2O2 at room temperature for 10 min. Subsequently, the slides were incubated in 3% pepsin solution at 37 °C for 20 min. After rinsing with PBS, they were pre-hybridized in pre-hybridization buffer at 37 °C for 3 h in a humid chamber. Hybridization was then carried out by adding 20 nM 5’-DIG-labeled probe diluted in hybridization buffer to the sections at 37 °C overnight. After a thorough wash with SSC, the sections were blocked at 37 °C (30 min), incubated with biotinylated mouse anti-DIG reagent at 37 °C (60 min), washed again, and incubated with streptavidin-biotin-peroxidase complex (SABC) at 37 °C (20 min), followed by three rinses with 0.5 M PBS. The sections were then incubated with avidin-biotin peroxidase complex at 37 °C (20 min) and washed again. The results were visualized after color developing using 3,3’-diaminobenzidine (DAB) and counterstaining with hematoxylin. Finally, the sections were dehydrated in a graded ethanol, cleared in xylene, and mounted with coverslips.
Evaluation of staining
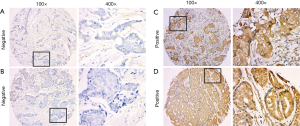
The expression of miR-361-5p was semi-quantitatively evaluated according to the intensity of the staining reaction. The intensity was graded as follows: 0—negative; 1—weak; 2—medium; and 3—strong. In this study, grade =3 was defined as miR-361-5p-positive staining, whereas grade ≤2 was defined as negative. The staining was assessed and scored separately by two independent observers in a blinded fashion.
Statistical analysis
The disease-free survival (DFS) was calculated from the date of surgery to the date of disease relapse. Event-free patients or patients without death were censored at the last follow-up. Survival curves were generated using the Kaplan-Meier method and compared by the Log-rank test. Using univariate and multivariate Cox regression analyses, the significance of various parameters for survival was analyzed. A two-sided P value <0.05 was considered statistically significant. All statistical analysis was performed using SPSS software (version 20.0; IBM Company).
Results
Patient characteristics
The clinical and pathological characteristics of the study cohort are presented in Table 1. The average age was 51.52 years (SD 10.19, median 50, range from 26 to 85). After a mean follow-up time of 96.57 months, 76 patients showed occurrence of disease events among the 375 cases.
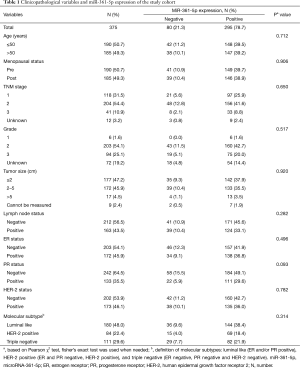
Full table
Expression pattern of miR-361-5p in breast cancer (BC) patients
In this cohort of 375 patients, TMAs were stained for miR-361-5p using ISH and representative images are shown in Figure 1 As shown in Table 1, positive miR-361-5p staining was observed in 78.7% (N=295; 78.7% positive, 21.3% negative) of the 375 cases according to the scoring criteria described above. Relationships between miR-361-5p expression and other parameters were analyzed using Chi-square tests. Clinicopathological variables like age, menopausal status, grade, tumor size and lymph node status were not significantly correlated with miR-361-5p expression (Table 1).
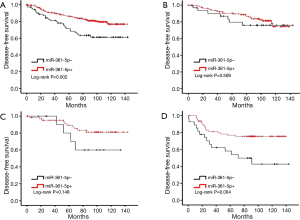
Elevated miR-361-5p expression is associated with better disease-free survival in breast cancer (BC)
To evaluate the clinical significance of miR-361-5p expression in BC, the relationship between the level of miR-361-5p expression and DFS was then analyzed. As the survival curves shown in Figure 2A, the clinical outcome of patients with positive miR-361-5p expression was significantly better than that of patients with negative miR-361-5p expression (P=0.002). The median DFS time was 95.52 months for positive miR-361-5p expression and 82.33 months for negative miR-361-5p expression.
In the univariate analysis (Table 2), the correlations between DFS and each clinicopathological parameter were examined. Several factors demonstrated a significant association with DFS: tumor size >5 cm (HR =3.108; 95% CI: 1.374–7.031; P=0.006), lymph node status (HR =1.979; 95% CI: 1.259–3.112; P=0.003), ER status (HR =0.615; 95% CI: 0.386–0.981; P=0.041) and PR status (HR =0.501; 95% CI: 0.292–0.860; P=0.012). And positive miR-361-5p expression indicated a lower risk for disease relapse (HR =0.471; 95% CI: 0.291–0.760; P=0.002).
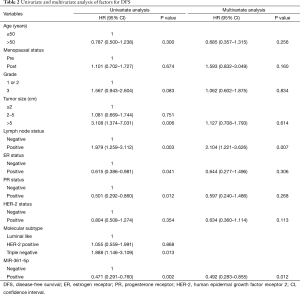
Full table
Then, in the further multivariate analysis (Table 2), only the lymph node status (HR =2.104; 95% CI: 1.221–3.626; P=0.007) and miR-361-5p expression (HR =0.492; 95% CI: 0.283–0.855; P=0.012) exhibited significant association. MiR-361-5p expression positively correlated with DFS in both univariate and multivariate analysis.
Moreover, the relationships between the level of miR-361-5p expression and survival according to different BC subtypes were also analyzed (Figure 2A-D). Among the three subtypes, the prognostic value of miR-361-5p was most significant among patients with TNBC for DFS (P=0.004). These results above indicated that miR-361-5p expression is an independent predictive factor for better prognosis in BC.
Discussion
BC is a heterogeneous disease with different morphological appearances, molecular features and behaviors (3). Many attempts have been made to identify BC signatures for diagnosis, prognosis, and prediction of the therapeutic response. Several studies have indicated specific miRNAs dysregulated in BC and they target multiple downstream genes and work in combination with each other, modulating one-third of all protein-coding genes (10,19). miRNAs play critical roles in responding to the current clinical needs, and could be used as easy, affordable, and clinically accessible molecular biomarkers in the retrospective analysis of large tissue collections and for diagnosis, prognosis, and prediction of therapeutic outcomes in BC (20).
MiR-361 is encoded on Xq21.2 in an intron between exon 9 and 10 of CHM/choroideremia (RAB escort protein 1), and gives rise to two mature miRNA species, miR-361-3p and the predominant miR-361-5p (21). It is reported that the target genes of miR-361-5p are VEGFA, MTUS1, RAD23B, MTR, GSR, TGFBR1, XRCC4, IL10, RB1, IGF1 and MTRR (22). Previous studies have indicated that miR-361-5p acts as a tumor suppressor in several cancers. In castration-resistant prostate cancer, miR-361-5p was demonstrated to be down-regulated through STAT/Bcl-xL pathway, suppressing cell proliferation and triggering apoptosis (16). While in colorectal carcinoma and gastric cancer, miR-361-5p also functions as a tumor-suppressive miRNA through binding to SND1 directly (15). And it was reported that miR-361-5p was inversely correlated with VEGFA expression in squamous cell carcinoma (22). However, a contradictory report identified that miR-361-5p acts as an oncogene in cervical cancer progression, enhancing cell proliferation and invasion (17). Besides, researchers have identified the most stably and highly expressed miRNA as candidate reference miRNAs in 14 types of common human tumors by the TCGA miRNA-seq data (23). And they demonstrated that miR-361-5p was the most recommended candidate reference miRNA in 9 of 14 human tumor types, including breast invasive carcinoma.
As far as we know, the role of miR-361-5p in BC has not yet been fully investigated and there is only one publication demonstrated that miR-361-5p up-regulated in metastatic BC (7 normal samples vs. 7 metastatic BCs) by screening with miRNA microarray (24). In the present study, we evaluated the expression of miR-361-5p by ISH analysis on TMAs and we found that miR-361-5p expression was positively associated with prolonged DFS. Moreover, both the univariate and multivariate analysis indicated that the expression of miR-361-5p was an independent prognostic factor for BC. Meanwhile, miR-361-5p density was remarkably associated with BC subtype. The prognostic value of miR-361-5p was most significant in patients with triple-negative breast cancer (TNBC). TNBC is defined as BC that lacks ER, PR, and HER2 expression. Compared with other subtypes of BC, TNBC shows a high rate of metastasis and poor prognosis (25). There are already several miRNA signatures that have been identified to correlate with TNBC clinical outcome (24). So it would be quite beneficial to be able to identify this subtype with more simple biomarkers or signatures.
The results were consistent with the findings of previous studies from other researchers that showed miR-361-5p functioned as a tumor suppressor. The study provided evidence that miR-361-5p was an independent marker of BC progression. However, the results are in contrast to the previous study that identified miR-361-5p to be up-regulated in metastatic BC. One of the possible reasons for the discrepancy could be the difference of sample size between the two studies. Another possible reason may be that the experiment methods were different. The majority of the published studies reporting the expression profiling of miRNA were performed using microarray technology or RT-PCR. In the present study, we evaluated the expression of miR-361-5p by ISH analysis. Compared with other methods, ISH can visualize microRNA expression at the cellular level, so it is the only method that can directly assess the actual distribution of miRNAs (26,27). ISH analysis can thereby provide more precise information.
There are several limitations of this study that should be noted. Firstly, the cohort was not very large and the number of excluded data due to ISH staining was a little high. And the number of recurrences was relatively low. Besides, the composition of the cohort was not representative of the BC population exactly. As comprehensive data on miR-361-5p expression are currently unavailable, further studies are needed to reveal the exact role of miR-361-5p in BC and BC subtypes in much larger populations. Further functional analyses are also needed to validate the possible utility of miR-361-5p.
In conclusion, this study provided evidence that miR-361-5p overexpression at diagnosis correlates with better DFS for BC patients. Our finding may therefore highlight the prognostic value and potential clinical application of miR-361-5p expression as a biomarker for BC, providing additional information for oncologists when individualizing cancer management.
Acknowledgements
Funding: The study was supported by grants from the National Natural Science Foundation of China (81202079, 81201531), the Shanghai Committee of Science and Technology Funds (11QA1401400, 12ZR1406200, 12DZ2260100 and 12140901502). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics Committee of FUSCC, and a written informed consent was signed by each patient.
References
- DeSantis CE, Fedewa SA, Goding Sauer A, et al. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin 2016;66:31-42. [Crossref] [PubMed]
- Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol 2014;15:e279-89. [Crossref] [PubMed]
- Polyak K. Heterogeneity in breast cancer. J Clin Invest 2011;121:3786-8. [Crossref] [PubMed]
- Wittner BS, Sgroi DC, Ryan PD, et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res 2008;14:2988-93. [Crossref] [PubMed]
- vat 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530-6. [Crossref] [PubMed]
- Dowsett M, Sestak I, Lopez-Knowles E, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol 2013;31:2783-90. [Crossref] [PubMed]
- Marrone M, Stewart A, Dotson WD. Clinical utility of gene-expression profiling in women with early breast cancer: an overview of systematic reviews. Genet Med 2015;17:519-32. [Crossref] [PubMed]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [Crossref] [PubMed]
- Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene 2006;25:6176-87. [Crossref] [PubMed]
- Graveel CR, Calderone HM, Westerhuis JJ, et al. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer (Dove Med Press) 2015;7:59-79. [PubMed]
- Iorio MV, Casalini P, Piovan C, et al. Breast cancer and microRNAs: therapeutic impact. Breast 2011;20 Suppl 3:S63-70. [Crossref] [PubMed]
- Tang Y, Zhou X, Ji J, et al. High expression levels of miR-21 and miR-210 predict unfavorable survival in breast cancer: a systemic review and meta-analysis. Int J Biol Markers 2015;30:e347-58. [Crossref] [PubMed]
- D'Aiuto F, Callari M, Dugo M, et al. miR-30e* is an independent subtype-specific prognostic marker in breast cancer. Br J Cancer 2015;113:290-8. [Crossref] [PubMed]
- Ma L, Li GZ, Wu ZS, et al. Prognostic significance of let-7b expression in breast cancer and correlation to its target gene of BSG expression. Med Oncol 2014;31:773. [Crossref] [PubMed]
- Ma F, Song H, Guo B, et al. MiR-361-5p inhibits colorectal and gastric cancer growth and metastasis by targeting staphylococcal nuclease domain containing-1. Oncotarget 2015;6:17404-16. [Crossref] [PubMed]
- Liu D, Tao T, Xu B, et al. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6). Biochem Biophys Res Commun 2014;445:151-6. [Crossref] [PubMed]
- Wu X, Xi X, Yan Q, et al. MicroRNA-361-5p facilitates cervical cancer progression through mediation of epithelial-to-mesenchymal transition. Med Oncol 2013;30:751. [Crossref] [PubMed]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc 2007;2:1508-14. [Crossref] [PubMed]
- Palmero EI, de Campos SG, Campos M, et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol 2011;34:363-70. [Crossref] [PubMed]
- Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015;5:1122-43. [Crossref] [PubMed]
- Afanasyeva EA, Hotz-Wagenblatt A, Glatting KH, et al. New miRNAs cloned from neuroblastoma. BMC Genomics 2008;9:52. [Crossref] [PubMed]
- Kanitz A, Imig J, Dziunycz PJ, et al. The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PLoS One 2012;7:e49568. [Crossref] [PubMed]
- Zhan C, Yan L, Wang L, et al. Identification of reference miRNAs in human tumors by TCGA miRNA-seq data. Biochem Biophys Res Commun 2014;453:375-8. [Crossref] [PubMed]
- Sun EH, Zhou Q, Liu KS, et al. Screening miRNAs related to different subtypes of breast cancer with miRNAs microarray. Eur Rev Med Pharmacol Sci 2014;18:2783-8. [PubMed]
- de Ruijter TC, Veeck J, de Hoon JP, et al. Characteristics of triple-negative breast cancer. J Cancer Res Clin Oncol 2011;137:183-92. [Crossref] [PubMed]
- Sempere LF. Tissue slide-based microRNA characterization of tumors: how detailed could diagnosis become for cancer medicine? Expert Rev Mol Diagn 2014;14:853-69. [Crossref] [PubMed]
- Zhang X, Lu X, Lopez-Berestein G, et al. In situ hybridization-based detection of microRNAs in human diseases. microRNA Diagn Ther 2013;1:12-23.


