Functional role of lncRNA DB327252 in lung cancer
Introduction
Lung cancer is still the most commonly diagnosed cancers among men, and the second among women nationwide in China; it is the foremost cause of death related to cancer in 2015 (1). Lung cancer involves a complex interaction of gene resulting in variable modulation of key pathways participate in tumor cell growth (2,3). Therefore, an improved study of the molecular mechanisms is needed for more efficient management related to lung cancer. For decades, the efforts are majorly focusing on the expression of protein-coding genes and their roles in the lung cancer pathogenesis (4,5). However, the majority of the human genome consists of long non-protein-coding RNA (lncRNA). LncRNAs may also act potential important players in the cancer phenotypes.
lncRNA is the RNA molecule has nucleotides longer than 200 and has no protein encoding capability (6). The role played by lncRNAs is still unknown but is being increasingly recognized (7,8). Unlike protein-coding genes, the lncRNAs transcripts’ functions and their relationships to diseases are mostly unclear (9,10). Such like some lncRNAs involve in carcinogenesis and progression of cancer (11,12). Therefore, the detail study of their functions and molecular mechanisms is extremely important to understand the process of tumors. Recent studies indicated that hundreds of lncRNAs were linked to lung cancer. These findings also raised the possibility that many of these lncRNAs are contributing to the lung cancer, including MEG3, H19 and SCAL1, etc. (13). However, the most part of the function of these lncRNAs is still unknown and the new cancer-related lncRNAs are waiting to be found.
DB327252 is a lncRNA that related to the lung cancer and is an important player in the phenotypes of the disease. Our study finds the high expression of lncRNA DB327252 in lung cancer cell lines and concludes that the expression has influences on the malignant process of lung cancer cell lines. Our outcomes demonstrated that the down-regulation of lncRNA DB327252 could suppress the lung cancer cells’ proliferation. These observations simply that lncRNA DB327252 was probable one of the oncogene genes, which provide new evidence that lncRNAs has its role in lung cancer.
Methods
Patient samples
Paired lung cancer and cancer-adjacent normal tissues were collected from 91 patients that had primary surgical resection of lung cancer in the Department of Thoracic Surgery, General Hospital of Guangzhou Military Command of Chinese People’s Liberation Army in Guangzhou, between January 2014 and June 2014. The cancer-adjacent normal tissues were collected 5 cm away from the edge of the tumor. Each freshly sample was immediately stored in liquid nitrogen until use after removed from the body. The extracted tumors were confirmed by pathological examination and staged based on the tumor-node-metastasis (TNM) staging system. Patients who participated in this study had not received radiotherapy, chemotherapy, targeted therapy, and/or immunotherapy treatment prior to the lung surgery. This clinical study has been approved by The Human Research Ethics Committee of General Hospital of Guangzhou Military Command of Chinese People’s Liberation Army and the consent has been accepted by all patients.
Cell lines
The human lung cancer cell lines A549 were acquired from the Sun Yat-sen University Cancer Center (Guangzhou, China). The cell line of 16HBE-T was donated by the Institute for Chemical Carcinogenesis of Guangzhou Medical University. This Institute laboratory transferred 16HBE cells to a malignant transformed cell line by induced with anti-BPDE (16HBE-T). The transform detail can be referred to the Dr. Jiang’s article (14). Both cell lines were retained in RPMI-1640 (Invitrogen, Carsbad, CA, USA) containing 10% inactivated fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin and 100 ug/mL streptomycin) at 37 °C in a humidified incubator with 5% (v/v) CO2.
Total RNA extraction
Total RNA was extracted from the frozen sample using TRIzol Reagent (Invitrogen) and then analyzed according to the manufacturer’s instruction; and the total RNA was dissolved by using 20 μL diethylpyrocarbonate-treated water.
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) analyses
The GoScript Reverse Transcription (RT) System (Promega, Madison, MI, USA) was used for generating cDNA. Based on the manufacturer’s instruction, we mixed well with the 2 µg total RNA, 1 µL oligo (dT) 15 primer, 1 µL random primer, 4 µL GoScript 5× reaction buffer, 2 µL MgCl2, 1 µL nucleotides mix, 0.5 µL recombinant RNasin ribonuclease inhibitor, 1 µL GoScript reverse transcriptase, and nuclease-free water. RT reaction was run for 1 hour at 42 °C. The expression of DB327252 was using GoTaq®qPCR master mix (Promega) on 7500 System (Life). All reactions were preformed following the manufacturer’s protocol.
siRNA transfection
Lung cancer cells were transfected with four different siRNA (siRNA-1, siRNA-2, siRNA-3, siRNA-4) that targeted lncRNA DB327252 and a scrambled siRNA control which were designed and synthesized by GenePharma (Shanghai, China) using Lipofectamine3000 (Invitrogen), based on the manufacturer’s instructions. The sequences of the four siRNAs and the scrambled siRNA were displayed in Table 1.

Full table
The DB327252 and controlled siRNAs were transfected into A549 and 16HBE-T cells. Cells were cultured in 6-well plate. And siRNA for common sequence and specific sequence of DB327252 were transfected respectively when the cells were at 50–60% confluency. The cells were collected 24 hours after transfection.
Determination of cell viability and colony formation assay
Transfected cells were examined their proliferation by using the Cell Counting Kit-8 (CCK-8; Dojindo, Tokyo, Japan). Cells (3×103) were put in 96-well plates with 100 μL of cell medium per well and cultivated for 24 hours under normal conditions. And then they were transfected with siRNAs. After 24, 48, 72 hours, 10 μL of CCK-8 reagent was injected in each well respectively. After incubated for 2 hours, A450 was inspected in each well using a Synergy 2 microplate reader (BioTek, Winooski, VT). The transfected and controlled cells were plated in a flesh six-well plates with media having 10% FBS, where medium will be replaced every 4 days. After 2 weeks, the colonies were fixed by methanol and dyed by 0.1% crystal violet for 30 minutes. More than 50 cells were counted manually. Triplicate wells were assessed for each treatment group.
Cell-cycle detection
After transfection, the cells were collected by trypsinization at 48 hours and were washed two times by using phosphate-buffered saline (PBS; Gibco, CA, USA). In order to performing flow-cytometric analysis of the cell cycle, the cells were tinted with propidium iodide by the Cycle testTM Plus DNA Reagent Kit (BD Biosciences), and finally analyzed immediately with flow cytometry (FACScan; Becton Dickinson, Franklin, NJ). The percentage of cells in the S, G0/G1, and G2/M phases were calculated and evaluated.
Lentiviral vector transfection
A549 and 16HBE-T were put in six-well plates 24 hours prior to transfection, and were nurtured with complete medium (2 mL) containing hexadimethrine bromide overnight at 37 °C with 5% (v/v) CO2. And then, cells were transfected by a mixture of 10 μL of virus diluted in 2 mL of complete medium with Polybrene (Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 5 μg/mL and were incubated with stable temperature at 37 °C with 5% (v/v) CO2 overnight. The medium was then changed with 2 mL of complete medium without Polybrene; and then the cells were continually cultured for another 48 hours. The efficiency of DB327252 inhibition was determined with qRT-PCR.
Tumor formation assay in a nude mouse model
Nude mice (4 weeks old) with Balb/c background were applied in this tumor formation assay. These mice were purchased from the Medical Animal Experimental Center of Guangdong Province. A549 and 16HBE-T were transfected with shRNAs or shRNAs-NC, which were suspended by PBS and washed twice. 2×107 cells/mL was injected on the right side of the posterior flank of each mouse. The tumor diameter was gauged every three days. After 4 weeks, the tumor xenografts were excised and weighed once they are removed from the mice. All of the surgeries were performed under sodium pentobarbital anesthesia.
Statistical analysis
SPSS version 16.0 (SPSS Inc., Chicago, IL, USA), as well as GraphPad Prism Software (GraphPad Software Inc., San Diego, CA, USA), were used for statistical analysis Values are stated in the format of means ± SD. The data were evaluated by applying statistical analysis methodology of Student’s t-test and one-way ANOVA; for all statistical analysis in this study, P<0.05 was considered statistically significant.
Results
Up-regulation of DB327252 in lung cancer tissues
A total of 91 paired clinical lung cancer tissues and cancer-adjacent normal tissues were evaluated for lncRNA DB327252 expression levels by applying qRT-PCR (Figure 1A). The mean expression level of DB327252 was 0.158±0.159 and 0.032±0.029 (DB327252/GAPDH ± standard deviation) in lung cancer tissues and non-tumorous tissues. The expression levels of DB327252 were appreciably higher in lung cancer tissues compared to cancer-adjacent normal tissues (P=0.000, Figure 1B,C).
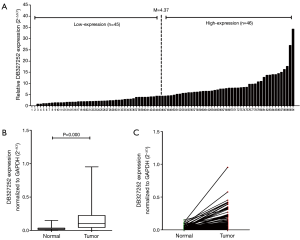
Association between the expression of DB327252 and the clinicopathological features in human lung cancer
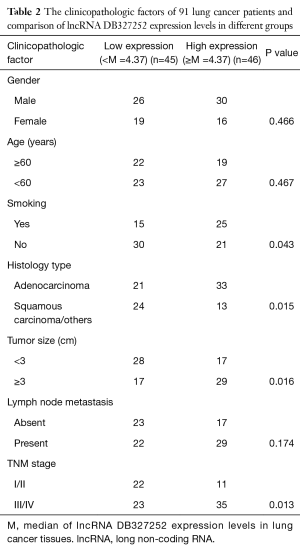
As the relationship between the DB327252 expression and clinical and pathologic features presented in Table 2, the expression level of DB327252 was no significantly related to gender, age, metastasis (P>0.05). The expression levels were statistically different between smoker and non-smoker group; however, this difference was considered as marginal as the P value is 0.043. A higher DB327252 expression level also have been observed under adenocarcinoma compared to squamous carcinoma/others (P=0.015). And the expression was also highly associated with advanced TNM stage (III/IV, P=0.013) and tumor size (≥3 cm, P=0.016); in addition, the higher level of expression was observed in advanced lung cancer stages.
Full table
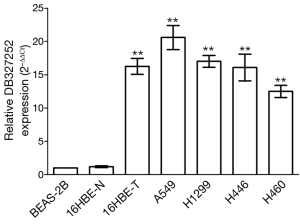
DB327252 expression in lung cancer cells
The analysis of qRT-PCR was used to measure the expression level of DB327252 in lung cancer cell lines. The result indicated that the expression level of DB327252 was at a relatively high level in five lung cancer cell lines, including 16HBE-T, A549, H1299, H446 and H460, compared to BEAS-2B or 16HBE-N (P=0.000, Figure 2).
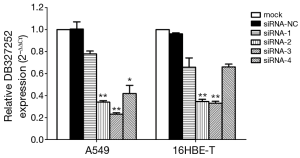
Knockdown of DB327252 expression by siRNA
We prepared four siRNAs (DB327252-1 to -4), which were stably transfected into 16HBE-T and A549 cells. DB327252 expression was markedly decreased, and DB327252-2 and DB327252-3 were more efficient than DB327252-1 and DB327252-4 in both 16HBE-T and A549 cells, which was detected by qRT-PCR. According to Quantification analysis, the result indicated that more than 60% of DB327252 expression was knocked down in siRNA group (Figure 3).
The effect of DB327252 on cell proliferation in the lung cancer cell lines
In order to identify the role of DB327252 in A549 and 16HBE-T cells, we preformed functional assays with A549 and 16HBE-T by knockdown the expression of DB327252 (DB327252/siRNA). In comparison of the controlled siRNA, transfection with DB327252/siRNA resulted in a considerable decreased viability in A549 and 16HBE-T. In addition, the data showed that the knockdown expression of DB327252 also largely reduced the colony-forming ability of the population (P<0.05, Figure 4A). A DNA content analysis with flow cytometry demonstrated that A549 cells transfected by siDB327252-2 and siDB327252-3 have a relatively higher cells proportion in G0/G1 phase, and had a reduced proportion in G2/M phase (P<0.01, Figure 4B). As expected, the growth inhibition was achieved as showing the cells’ proportion and was reduced in the G2/M phase while the cells’ proportion was appear raised in G0/G1 phase as transfecting 16HBE-T cells with siDB327252-2 and siDB327252-3 (P<0.05, Figure 4C). These results indicate that in response to DB327252 down-regulation, the increase of proportion in both cells lines were both occurred in GO/G1 phase. Results of these experiments demonstrated that the down-regulation on DB327252 expression caused the increase of the G0/G1 stage cells, which lead to the inhibition of cell growth.

Stable knockdown of DB327252 expression
We found a stably transfected cell line which can persistently constrict the expression level of DB327252 for additional functional analysis, because DB327252 expression can be inhibited by siRNAs for only a few times. The lentivirus-mediated shRNAs or shRNA-NC was transfected into A549 and 16HBE-T cells. As they mixed the Green Florescent Protein (GFP) together with the shRNAs, we can detect the GFP-positive cells just like the green florescent. The level of DB327252 would be detected by qRT-PCR, as it indicated in Figure 5A, the expression level of DB327252 was notably reduced by transfected of DB327252 shRNAs compared to the control group (P<0.01).
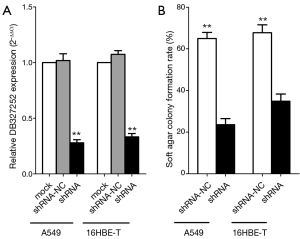
Soft agar colony formation assay
In order to elucidate the functions of DB327252, the soft agar assay was applied in the independent growth of lung cancer cells. As revealed in Figure 5B, a dramatically decrease in the colony-forming efficiency has been observed after transfected with shRNAs compared to shRNA-NC transfected group. The down-regulation of DB327252 restrained the growth capacity of A549 and 16HBE-T cells. Moreover, the transfected of DB327252 shRNAs mildly suppressed colony forming ability compared to the shRNA-NC (P<0.05).
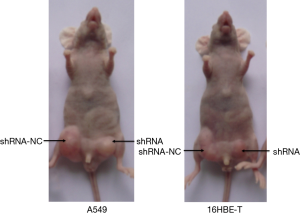
Knockdown of DB327252 expression suppressed lung tumor growth in vivo
Although DB327252 function was illustrated in two cell lines, there was only little evidence to support in vivo. In order to provide the DB327252 effect in lung cancer, 4-week-old Balb/c nude mouse model was used. The subcutaneous injection with transfected or controlled cells was performed in the mice of each group. After injection, all mice developed tumors in one week. In comparison from the mice tumor, the tumors caused by DB327252-shRNA-transfected cells injection were apparently smaller (Figure 6A,B) and lighter (Figures 6C,7) than those derived from the shRNA-NC-transfected cells injection. These results clearly showed that the DB327252 suppression can inhibit the growth of tumor.

Discussion
The lncRNAs’ functional mechanism is the most difficult and least known aspect in lncRNA research (15,16). Recent studies have exposed that a significant amount of lncRNAs are transcribed from the human genome, and have been newly attributed expansive roles compared to the previous studies across various biological processes. Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT-1) is one of the most classic oncogenic lncRNA. This lncRNA has been reported widely express in human organs and act as an independent prognostic factor as a probable biomarker for metastasis and survival rate for patient with hepatocellular carcinoma (HCC) and non-small cell lung cancer (NSCLC) (17-19). MALAT-1 is found localized on nuclear speckles, which is known containing several proteins involved in alternative splicing (20). MALTA-1 is found to structure a molecular scaffold to modify numerous proteins found on nuclear speckles, and to adjust the SR proteins’ phosphorylation. Exhausting MALAT-1 can modify the configuration of alternative splicing of an mRNA subset.
Another example is HOX transcript antisense intergenic RNA (HOTAIR). HOTAIR was found having a relatively low expression level in normal breast tissue while having a higher level in breast cancer tissue. As the study reported, HOTAIR expression is related to a poor prognosis in cancer disease (21,22). By comparing the 42 paired NSCLC tissues and cancer-adjacent normal tissues in 2013, Dr. Liu found that the HOTAIR expression level in cancer tissue is notably advanced compared to the cancer-adjacent normal tissue (23). The expression of HOTAIR is correlated to the TNM stage, the level of lymph node metastasis, and prognosis. The further experiment on nude mice also shows that the number of lung metastases was notably reduced through the knockdown of HOTAIR expression. Some studies found that the up-regulation of HOTAIR, by down-regulating the Homeobox protein Hox-A5 (HOXA5) and up-regulating the Matrix metalloproteinases (MMPs), promoted the invasion and metastasis capability of NSCLC. HOXA5 was majorly found in alveolar epithelial cells, and it has a pivotal role in the lung growth. The interference on HOXA5 expression leads to the mice lung’s abnormal development, form, and function; however, the detail regulatory mechanism of HOXA5 is still unclear. MMPs participate in the degradation and remodeling of extracellular matrix (ECM) and basement membranes, which is the main steps for the tumor growth and metastasis. Therefore, the up-regulation of HOTAIR, and overexpression of MMPs that promote degradation of ECM and basement membranes, is in favor of the invasion and metastasis of NSCLC (24).
By activating tumor suppressor pathways, tumor-suppressor lncRNAs could affect cells and their function. To support this point, there are several examples. First of all, lncRNA-P21 plays an important role in p53 pathway and participates in cell apoptosis (25). Another lncRNA is Growth Arrest-Specific 5 (GAS5). GAS5 was down-regulated in various types of tumor, such as breast and prostate cancers. Perhaps it is affected apoptosis by monitoring activities of glucocorticoids (26,27). These studies provide a new avenue for cancer research, by which we could understand carcinogenesis more in-depth; and lncRNA DB327252 has also been newly identified in relation to abnormal expression in lung cancer in our study. However, the mechanism of DB327252 in the development of lung cancer remains unidentified.
In this study, we confirmed that lncRNA DB327252 is over-expressed in lung cancer tissues compared to cancer-adjacent normal tissues. We also revealed that lncRNA DB327252 is highly expressed in lung cancer cell lines in comparison with human bronchial epithelial cell lines. Additionally, we evaluated the expression level of a long noncoding RNA DB327252 in lung cancer tissue and cancer-adjacent normal tissue. Results indicated that lncRNA DB327252 was significantly up-regulated and correlated with smoking, histology type, tumor size, and TNM stage.
The expression of lncRNA DB327252 in lung adenocarcinoma is significantly higher than in lung squamous cell carcinomas/others. This observation is also implies that DB327252 can become a potential biomarker for the histological type. We found lncRNA DB327252 expression was higher in ≥3 cm tumor size and advanced TNM stage (III/IV) than in <3 cm tumor size and earlier TNM stage (I/II). This indicated that lncRNA DB327252 might function as an oncogene in lung cancer tumorigenesis. As high expression level of lncRNA DB327252 was related to an aggressive tumor phenotype in lung cancer, we considered that lncRNA DB327252 could be a significant part in tumor biology. We designed to synthesize several siRNAs against lncRNA DB327252 and oligonucleotides with negative control, then transfected them into A549 and 16HBE-T cells, and then performed the qRT-PCR assays analysis to try to uncover the most effective siRNA sequence and to avoid the off-target. Our findings indicated that the knockdown of lncRNA DB327252 restrained cell proliferation, and led to cell cycle arrested at the G1/G0 phase.
As the well-established method, the soft agar colony formation assay used to characterize the capability in vitro, which method is also considered as one of the most stringent tests for cells malignant transformation (28). The soft agar colony formation assay is also applied trying to discover inhibitors of tumorigenicity in lung cancer cells (29). It is widely accepted that, the capability of cancer cells forming colonies in agar is related to their tumorigenicity. The experiment in vitro also verified that knockdown of DB327252 reduced the expression level and inhibited the colony formation abilities. Moreover, in vivo experiment confirmed that the knockdown of DB327252 inhibited tumorigenic ability of A549 and 16HBE-T cells in nude mice.
Based on our research, this is the first report showed that lncRNA DB327252 had different expression level between lung cancer tissues and adjacent normal tissues. Moreover, this is the first time to prove lncRNA DB327252 could regulate the proliferation of lung cancer cell as well. We showed that lncRNA DB327252 was significantly over-expressed in lung cancer tissues and regulated lung cancer cell proliferation both in vitro and in vivo. Although lncRNAs were identified to have play a significant biological roles in human diseases, regulating the expression level of lncRNAs remain largely unknown. Our result demonstrated that lncRNA DB327252 is related to lung cancer and has a significant role in both A549 and 16HBE-T cancer cells. This study implies that lncRNA DB327252 has an oncogene-like function in lung cancer. In summary, the down-regulation of DB327252 via siRNA reduced the expression levels of DB327252, and inhibited the proliferation and tumorigenicity abilities of lung carcinoma cells. These findings expand our knowledge of lncRNA DB327252, and support its oncogene-like function in lung cancer.
Acknowledgements
Funding: This work was supported by the Medical scientific research funds of Guangdong (B2011257), Medical funds of Jieping Wu (320.6750.12709) and Science and Technology Program of Guangzhou, China (201607010117).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This clinical study has been approved by The Human Research Ethics Committee of General Hospital of Guangzhou Military Command of Chinese People’s Liberation Army and the consent has been accepted by all patients.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [Crossref] [PubMed]
- Zhou Q, Zhang XC, Chen ZH, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3316-21. [Crossref] [PubMed]
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet 2004;5:316-23. [Crossref] [PubMed]
- Johnson JL, Pillai S, Chellappan SP. Genetic and biochemical alterations in non-small cell lung cancer. Biochem Res Int 2012;2012:940405.
- Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012;31:4577-87. [Crossref] [PubMed]
- Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 2012;279:3159-65. [Crossref] [PubMed]
- Zhou H, Xiao B, Zhou F, et al. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers 2012;17:104-10. [Crossref] [PubMed]
- Cabianca DS, Casa V, Gabellini D. A novel molecular mechanism in human genetic disease: a DNA repeat-derived lncRNA. RNA Biol 2012;9:1211-7. [Crossref] [PubMed]
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev 2013;22:2240-53. [Crossref] [PubMed]
- Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013;34:613-20. [Crossref] [PubMed]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38. [Crossref] [PubMed]
- Hu G, Yang T, Zheng J, et al. Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Mol Carcinog 2015;54 Suppl 1:E192-204. [Crossref] [PubMed]
- Jiang Y, Chen J, Chen X. Malignant transformation of human bronchial epithelial cells induced by benzo(a)pyrene metabolite dihydroxyepoxy benzo pyrene. Wei Sheng Yan Jiu 2001;30:129-31. [PubMed]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet 2009;5:e1000459. [Crossref] [PubMed]
- Yasuda J, Hayashizaki Y. The RNA continent. Adv Cancer Res 2008;99:77-112. [Crossref] [PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. [Crossref] [PubMed]
- Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [Crossref] [PubMed]
- Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 2011;6:1984-92. [Crossref] [PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013;13:464. [Crossref] [PubMed]
- Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep 2012;45:604-11. [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8. [Crossref] [PubMed]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009;28:195-208. [Crossref] [PubMed]
- Borowicz S, Van Scoyk M, Avasarala S, et al. The soft agar colony formation assay. J Vis Exp 2014.e51998. [PubMed]
- Horibata S, Vo TV, Subramanian V, et al. Utilization of the Soft Agar Colony Formation Assay to Identify Inhibitors of Tumorigenicity in Breast Cancer Cells. J Vis Exp 2015.e52727. [PubMed]


