
Potential treatment strategy for the rare osimertinib resistant mutation EGFR L718Q
Introduction
The incidence of mutations in the epidermal growth factor receptor (EGFR) gene is about 50% in non-small cell lung cancer (NSCLC) patients in China (1). In the era of rapidly developing targeted therapies, the prognosis of advanced NSCLC has significantly improved, owing to the wide use of EGFR tyrosine kinase inhibitors (TKIs). Meanwhile, the emergence of de novo or secondary resistant mutations is a great clinical challenge. One of the novel resistant mutations, EGFR L718 substitutions (with the L718Q mutation being dominant clone), was reported to be identified in 8% of the osimertinib-resistant Chinese NSCLC patients (2). It has been revealed that EGFR L718Q independently leads to osimertinib resistance by stabilizing its non-reactive conformation (3). However, no report has yet identified how to effectively treat this rare mutation.
Case presentation
The patient was a 44-year-old Asian man who had no history of smoking. He was admitted to a local hospital because of the chest pain and dry cough on March 17, 2016. The chest computed tomography (CT) showed a mass (measuring 10 mm in diameter) in the posterior segment of the left upper lobe (LUL), together with metastatic nodules in the bilateral lungs and the pleura (Figure 1A). The left subcarinal lymph nodes were enlarged. The pleural effusion pathology identified atypical cells. CT-guided LUL mass biopsy revealed invasive lung adenocarcinoma (acinar predominant). Immunohistochemistry (IHC) indicated the TTF-1 (+) and ALK(D5F3) (−). The EGFR L858R mutation was detected in the biopsy sample by the qualitative amplification-refractory mutation system (ARMS)-PCR, and also detected in the next-generation sequencing (NGS)-based ctDNA genetic testing (with a frequency of 0.65%). Briefly, the ctDNA was obtained using QIAamp Circulating Nucleic Acid kit (Qiagen) from plasma and quantified using Qubit 2.0 Fluorimeter (Life Technology), followed by standardized NGS library preparation and sequencing on Nextseq500 sequencer (Illumina) (4). Abdominal and brain magnetic resonance imaging (MRI) with contrast, and a bone scan were performed, and distant metastases were ruled out. The final diagnosis was stage IV lung adenocarcinoma (C-T4N2M1a), with EGFR L858R mutation.
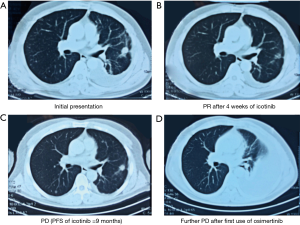
Because the EGFR-targeted therapy was temporarily unavailable in the patient’s area, beginning March 2016, the patient received 4 courses (16 weeks) of cisplatin/pemetrexed, and remained stable disease (SD) according to the response evaluation criteria in solid tumors (RECIST) criteria. Icotinib (oral) was initiated in June 2016. After 4 weeks of icotinib treatment, chest CT confirmed significant shrinkage of the LUL mass and the mediastinal lymph nodes, and some of the bilateral metastatic nodules disappeared. The patient achieved a partial response (PR) (Figure 1B). However, in the follow-up CT scans, which were performed every 4 weeks, a gradual increase in the size and density of the bilateral nodules were reported. As he remained asymptomatic, the patient continued the icotinib treatment for another 32 weeks. In March 2017, the chest CT demonstrated that the LUL mass increased to 14 mm in diameter. Increased number of intrapulmonary and pleural metastases were also present, with the largest one in the left lower lobe (LLL) measuring 16 mm in diameter. Progression of disease (PD) was confirmed after 9 months of icotinib treatment (Figure 1C). At the same time, the resistant mutation, EGFR T790M was detected by NGS-based ctDNA genetic testing, and osimertinib was immediately initiated. After 4 weeks of osimertinib treatment, chest CT showed further enlarged metastatic nodules, emergence of a large volume of pleural effusion, and atelectasis, indicating a further PD (Figure 1D). The patient was referred to this hospital.
Considering that the patient's response to osimertinib was poor, although there was no other significant findings in the ctDNA-based genetic testing, it could not be ruled out that there was a key resistant mutation which had not yet been captured by the ctDNA tests. It was necessary to perform comprehensive NGS-based genetic profiling on the tissue specimen. Since the patient’s initial biopsy specimen had been exhausted, and there was no nodule which was available through another biopsy, he received a left lower lobe wedge resection, and a metastatic lesion measuring about 10 mm was obtained. A minimal use of tissue for IHC revealed the consistent phenotypes, and the formalin-fixed, paraffin-embedded tissue was then subjected to the NGS-based genetic testing of 168 lung cancer-related genes. The quantitative analysis estimated that the EGFR L718Q mutation frequency was 68.84%, and the EGFR L858R mutation frequency was 89.7%. The EGFR amplification with a copy number (CN) of 11.54 was also detected. The EGFR T790M mutation was negative in this resection specimen.
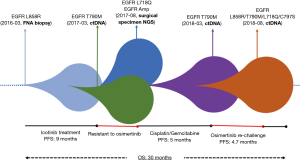
The EGFR L718Q mutation was first reported in 2016 (5). It was identified in a NSCLC patient with acquired osimertinib resistance. In the following in vitro studies, it has been shown to independently lead to first and third-generation TKI resistance (6). Besides EGFR L718Q, a EGFR amplification with a CN of 11.54 that led to broad-spectrum TKI resistance, was also present in this patient. He therefore would not benefit from any targeted therapy (7). In combination with that the previous conventional chemotherapy regimen of cisplatin/pemetrexed was not effective, after a pharmacogenomics evaluation, a personalized chemotherapy regimen of cisplatin/gemcitabine was chosen for this patient and initiated in October 2017. After 1 course of the chemotherapy, his bilateral nodules shrank markedly, and a PR was observed. In March 2018, the patient developed PD after 4 courses of chemotherapy. Chest CT showed increasing number of multiple lung nodules, with some being enlarged. Multiple distant metastases were found in the brain. At the same time, the EGFR T790M mutation was identified again in ctDNA testing with a frequency of 0.18%. Osimertinib was immediately given to re-challenge. The following chest CT after 4 weeks of osimertinib treatment indicated obvious decrease in the number of the lung nodules, and significant shrinkage was observed in both the lung nodules and brain metastases. The patient achieved PR. The progression-free survival (PFS) of this osimertinib re-challenge was 4.7 months until August 17, 2018, when the bilateral lung nodules and brain metastases were enlarged. Another osimertinib-resistant mutation, EGFR C797S, was detected in the ctDNA with a frequency of 0.60%, which co-existed with the EGFR L858R mutation at the frequency of 6.78%, EGFR L718Q mutation at the frequency of 3.32%, and EGFR T790M mutation at the frequency of 0.52%. The patient suffered from a great symptom burden of brain metastasis and refused any further targeted or systemic therapy. Supportive treatments were provided, and he died on September 9, 2018. His overall survival (OS) was 30.2 months (Figure 2).
iMDT discussion
Department of Thoracic Surgery
In this case, a stage IV lung adenocarcinoma (C-T4N2M1a) patient initially presented with a sensitizing EGFR mutation, EGFR L858R, and the PFS of the icotinib treatment was 9 months. However, he did not respond to the subsequent third-generation TKI treatment at all when the PD and the corresponding EGFR T790M mutation was indicated. It was hypothesized that there was a potential resistant mutation not detected in either the biopsy sample or the liquid biopsy. Through a comprehensive NGS approach on a resection specimen, he was finally found to have the rare EGFR L718Q mutation at a frequency of 68.84%, in addition to the EGFR amplification at a copy number of 11.54. His poor response to the initial osimertinib treatment was finally retrospectively explained. Although there is a view that liquid biopsy testing is capable of representing the comprehensive genetic profile of advanced patients (8), this case strongly suggests that in select patients, comprehensive genetic profiling can only be achieved with the NGS approach on a resection specimen obtained from the appropriate surgery.
Department of Molecular Oncology
In this case, it was hard to confirm whether the EGFR L718Q mutation was a de novo or secondary mutation. However, considering that EGFR L718Q leads to both first and third-generation TKI resistance, and the patient responded to the initial icotinib treatment, the acquired resistance to icotinib and osimertinib was more likely due to a secondary EGFR L718Q mutation.
When the resistant EGFR L718Q mutation and EGFR amplification were confirmed, it was clear that the patient would not continue to benefit from conventional targeted therapy. We indicated that for a patient with the rare EGFR L718Q mutation, a personalized chemotherapy would effectively “clear” the composition of the heterogeneous tumor mass. The patient significantly benefited from the osimertinib re-challenge when the sensitive cells were dominant. Chemotherapy could be a crucial component of the precision medicine, and significantly changed the patient’s response to the targeted therapies.
Moreover, we simultaneously observed the EGFR L718Q and the EGFR amplification when the patient was resistant to the osimertinib treatment, and identified four EGFR mutations in his late stage: EGFR L858R, T790M, L718Q, and C797S. The coexistence of these EGFR mutations suggested potential a TKI resistance mechanism which required further exploration.
The following issues regarding the diagnosis and treatment of this patient were discussed further
Question 1: As the specificity and sensitivity of ctDNA in advanced lung cancer patients are still under debate, is it still too early to regard this minimally invasive technique as an alternative for identifying all potentially actionable gene mutations?
Expert opinion 1: Dr Elisabetta Rossi and Dr. Rita Zamarchi
In principle, the superiority of liquid biopsy derives from the sample source, since peripheral blood could provide the genetic landscape of all cancerous lesions at any time, detecting genomic alterations responsive to a specific therapy or associated with drug resistance. However, the validity of this assumption depends on the sensitivity and specificity of the tests we use for genetic profiling; indeed, the clinical case discussed here support this view.
Generally speaking, we should consider some recommendations in using liquid biopsy.
First, concerning ctDNA, we should analyze a panel of actionable genes—it is limiting to test one mutation at once—and we should plan to re-analyze them during treatment, in order to reveal changes induced by the therapy as early as possible.
Second, nowadays liquid biopsy represents a class of biomarkers—ctDNA, circulating tumor cells (CTCs), tumor derived extracellular vesicles (tdEVs)—whose complementary use is recommended, in order to obtain more information from the same sample (9). For example, ctDNA can be used to detect actionable mutations, whilst CTCs are useful to predict treatment effectiveness (10). Moreover, concerning molecular analyses, the volume increase of analyzed samples should allow addressing the sensitivity limit, at least for CTCs (11).
We think that the technology is mature to investigate in prospective trials, whether this diagnostic/monitoring strategy satisfies criteria required for becoming standard of care, i.e., it offers time and quality of life to the patients, at lower cost for the National Health Systems (12).
Expert opinion 2: Dr. Marc G. Denis
The possibility of identifying molecular alterations without having to biopsy patients is extremely attractive. Numerous studies have shown that this approach can be very productive. The techniques used are more and more sensitive. In some cases, this can be done at the expense of specificity. This specificity must therefore be mastered in order to know whether the detected alterations are indeed present in the patient’s tumor. For instance, a recent study evaluated four NGS gene panel assays for mutations in ctDNA using replicate sets of 24 plasma samples. The authors identified a significant number of false-negative and false-positive variants and revealed substantial variability among the ctDNA assays (13). It is therefore necessary to use perfectly validated techniques by carrying out prospective studies. Only in this case will it be possible to use the results of circulating DNA tests to guide the choice of treatment.
ctDNA analysis has the advantage of being less sensitive to tumor heterogeneity. Indeed, the alterations present in all tumor sites can theoretically be found in one plasma sample. Conversely, all the molecular alterations found in a DNA sample are not necessarily present in the same tumor or even in the same cell. This may explain why in some cases tumor sites do not shrink during targeted therapy, although the corresponding molecular alteration has been identified in ctDNA.
Expert opinion 3: Dr. Carlos Camps and Dr. Amaya B. Fernandez-Diaz
Liquid biopsy analyzing ctDNA represents one of the most advanced investigation fields in clinical practice for lung cancer patients, and there are two scenarios where liquid biopsy plays a crucial role: the initial molecular diagnosis and assessment during targeted therapy. Although questions regarding sensitivity and clinical utility are still under debate, this technique presents several advantages over tissue biopsy (14).
This non-invasive technique allows performing the molecular diagnosis in patients with suboptimal clinical condition or with unfavorable tumor site to perform tissue biopsy, reducing the risk of major complications. Additionally, analysis of ctDNA allows performing all the required analysis, while scarcity amount of tissue can be limiting. And what is more important in our opinion, in opposition to single biopsies ctDNA has the capability to reflect the systemic tumor burden, intratumoral heterogeneity among the primary and metastatic lesions, and the capability to monitor molecular cancer evolution during the course of therapy (14,15).
According to NSCLC guidelines, all patients whose molecular status should be investigated are eligible for molecular determination in ctDNA. Lack of standardization is limiting implementation of ctDNA as an alternative technique to tissue biopsy when this one is available, however liquid biopsy has significant potential to improve patients care and implementation in clinical practise is necessary.
Several analytical methods have been developed for molecular assessment using ctDNA differing in accuracy, time to results and costs. But in general, the Achilles heel for ctDNA analysis is sensitivity despite its great specificity. Sensitivity depends on the ability to detect ctDNA, considering that ctDNA levels represents a small proportion of total circulating cell-free DNA (cfDNA), varying from less than 0.1% to over 10% depending on disease burden, stage, cellular turnover, and treatment response (16). Therefore, standardization of preanalytical conditions and good choice of the analytical technique play a crucial role to overcome the challenge.
From all analytical methods, broad NGS assays are the best alternative to detect all potentially actionable mutations by liquid biopsy and are preferred if available. NGS assays present acceptable levels of sensitivity of approximately 85%, efficient use of limited DNA with wider variety of genetic alterations detected simultaneously. Costs are progressively reducing.
Different NGS-based methods have been already validated for NSCLC ctDNA mutation detection, and there are numerous studies aimed to confer clinical validation and standardization of ctDNA analysis at diagnosis setting and progression to targeted therapy.
C. Camps laboratory working group has participated in the design and sample analysis of a multi-institutional prospective study including consecutive EGFR, ALK, ROS-1-altered NSCLC patients with TKI resistance from 12 Spanish institutions, performing ctDNA NGS by Guardant360 assay to impact in the clinical care of patients. In the study 64% of patients showed reliable evidence of tumor-DNA shed for resistance assessment and 24% of patients had actionable alterations; 17% of patients received molecular-guided therapies indicated by plasma NGS alone, and 4% needed plasma and tissue NGS sequencing (17).
RING project, is another multi-institutional prospective study conducted by GECP (C. Camps’s group) trying to evaluate the agreement of methodologies available for T790M testing in liquid biopsies in Spain, based on lack of comparison across different platforms to improve patient management (18).
Currently liquid biopsy is repetition considered a solid complement to tissue biopsy and the best alternative when the last one is not available. However, this diagnosis technique is being more used gradually, and in the near future when balance between accuracy and costs is achieved, will be an implemented alternative to tissue biopsy in clinical practice.
Question 2: For patients resistant to EGFR-targeted therapies, how can the timing of systemic chemotherapy and that of a subsequent targeted therapy re-challenge be precisely determined?
Expert opinion 1: Dr Elisabetta Rossi and Dr. Rita Zamarchi
We think that the complementary use of CTCs and ctDNA might allow monitoring loss of effectiveness of systemic therapy—when the CTCs result higher than the cut-off value of 3 cells (19)—in parallel disclosing drug resistance (20) or actionable mutations, at ctDNA level, as in the case discussed here.
Expert opinion 2: Dr. Marc G. Denis
It is generally accepted that the change in therapy should take place when the patient is no longer taking benefit from treatment. Several studies have in fact demonstrated that it was interesting to pursue the treatment beyond radiological progression. The issue of re-challenge with targeted therapy should be studied as part of a prospective clinical trial.
Expert opinion 3: Dr. Carlos Camps and Dr. Amaya B. Fernandez-Diaz
Some studies have assessed the clinical benefit of re-administering first- and second-generation EGFR-TKI after initial benefit to EGFR-TKI treatment and progression to second line platinum-based chemotherapy, showing moderate activity with a median PFS of approximately 3 months (21,22). However, the benefit of Osimertinib rechallenge is still uncertain.
The rationale behind rechallenging with an EGFR-TKI after intervening chemotherapy is based on the consideration that chemotherapy may eradicate the clones of cancer cells that are responsible for clinical resistance to a given EGFR-TKI, and regrowth of EGFR-TKI-sensitive cells can occur, which may re-sensitize the tumor to the inhibitor.
Therefore, the most important factor to propose EGFR-TKI re-challenge depends on the tumor molecular profile after intervening chemotherapy. NGS platforms are able to assess the presence of sensitizing mutations for a specific TKI such as T790M for Osimertinib, and other competitive mechanisms of resistance, these acquired resistance mechanisms can be EGFR related or EGFR non-related, while tertiary EGFR mutations are present in around 9% (the most common C797S in 20–30% of cases), EGFR-independent resistance mechanisms to Osimertinib such as PIK3CA, KRAS, BRAF, HER-2 and MET amplifications comprise around 50% of Osimertinib resistance mechanisms (23,24).
The utility of liquid biopsies analyzing ctDNA to monitor EGFR gene and detect mechanisms of primary and acquired resistance to treatments, has been extensively investigated to direct additional lines of therapies and to guide rechallenge protocols, considering that single tumor biopsy may fail to reflect tumor heterogeneity (25).
In this scenario, due to lack of standard of care after T790M-targeting third-generation EGFR-TKI progression, rechallenge with a specific EGFR-TKI after intervening chemotherapy might be a reasonable option to prolong overall survival and improve quality of life for selected patients (26). However, patient selection must be assessed by molecular analysis, detecting possible resistant mechanisms avoiding ineffective treatments.
Question 3: How should a personalized chemotherapy regimen for the patients resistant to conventional first-line cisplatin/pemetrexed be selected?
Expert opinion 1: Dr Elisabetta Rossi and Dr. Rita Zamarchi
Based on the case discussed here, at the onset of resistance to standard first-line chemotherapy, a personalized chemo regimen should be planned, if the NGS analysis of ctDNA do not reveal actionable mutations.
Beyond to recommend a more comprehensive investigation of the circulating compartment—including CTCs—the validity of this approach should be proved in prospective studies, before becoming standard of care. Therefore, we should consider exploiting the strategy of N-of-1 trial that has recently received great attention to speed up clinical research on the new frontier of precision oncology (27).
Expert opinion 2: Dr. Carlos Camps and Dr. Amaya B. Fernandez-Diaz
Platinum-based chemotherapy is the standard of care in advanced NSCLC for patients who are not suitable for targeted therapies or immunotherapy. However, the therapeutic efficacy of platinum-based chemotherapy varies remarkably among different individuals, with a response rate from 26% to 60% in NSCLC.
Pharmacogenomic studies try to explain the genetic bases for interindividual differences to predict the safety, toxicity, and/or efficacy of drugs. Great efforts have been made to try to identify patient´s genetic characteristics to predict therapeutic effect, not just to cisplatine, but others chemotherapeutic agents for NSCLC (28-30). For example, according to the mechanism of platinum, important components of DNA repair pathways (ERCC1, XPD, XPG, XRCC1 and XRCC3) have been related with the efficacy of platinum-based treatment and clinical outcome for NSCLC patients. However, published data shows insufficient evidence to identify molecular predictive markers for chemotherapy in lung cancer patients (30,31).
Therefore, chemotherapy treatment selection in NSCLC is currently based on anatomopathological and clinical criteria. Considering performance status, comorbidities, previous lines of treatment, or contraindications to certain type of treatment as is the case of antiangiogenic drugs, to help clinicians in the therapeutic decision (32).
Question 4: Is there a possible mechanism determining the presence of the quadruple EGFR mutations (EGFR L858R/T790M/L718Q/C797S)?
Expert opinion 1: Dr Elisabetta Rossi and Dr. Rita Zamarchi
In the case discussed here, the simpler explanation of the four mutations is that they reflect tumor heterogeneity. Indeed, the molecular analyses were performed on ctDNA, not in CTCs at single-cell level. Hence, we cannot exclude that the four mutations documented in this patient, derived from different subset of tumor cells that emerged in metastatic lesions after prolonged treatment.
Expert opinion 2: Dr. Marc G. Denis
Cancer cells acquire resistance to systemic treatment as a result of clonal evolution and selection. This is the case, for example, for the various alterations in the EGFR gene described in patients progressing on EGFR TKIs. The T790M mutation is a well-described mechanism of resistance to 1st and 2nd generation inhibitors. The C797S mutation is a mechanism of resistance to osimertinib. The co-existence of 3 mutations (activating mutation + T790M + C797S) has therefore been described in many patients treated with different inhibitors. The existence of a 4th mutation (L718Q) is described in this patient. This is possible in theory, even if in this patient the analysis, carried out on circulating DNA, does not make it possible to know whether these different alterations are present in the same cell, or even in the same tumor site.
Expert opinion 3: Dr. Carlos Camps and Dr. Amaya B. Fernandez-Diaz
Deletions in exon 19 and L858R missense substitutions are the most common druggable mutations in NSCLC, despite initial benefit of first- and second-generation EGFR-TKI progression is unavoidable with median time of 9-14 months. Drug resistance due to T790M detected in more than 50%, is the most common resistance mechanism to first- and second-generation EGFR-inhibitors. Nonetheless, other mechanisms have been described in smaller proportion, including HER2 and MET amplifications, histologic transformation and PIK3CA and BRAF mutations (33).
T790M-targeting third-generation EGFR-TKIs can overcome this secondary resistance, but the appearance of tertiary resistances is unavoidable as well due to different mechanisms previously described. It is known that the appearance of EGFR mutations in C797S and in smaller proportion L718Q confers resistance to osimertinib (34). These EGFR tertiary mutations have been already reported in metastatic NSCLC coexisting with EGFR L858R and T790M mutations in patients who progressed on osimertinib treatment (5,35).
The presence of multiple EGFR mutations concurrently can be explained by the association of tumor heterogeneity and dynamic changes induced by sequential EGFR-TKI treatment, increasing genomic complexity.
NSCLC is a heterogeneous disease which harbours co-occurring genomic alterations, even NSCLCs driven by a dominant oncogenic alteration show intradriver molecular diversity, what is translated into heterogeneous clinical behaviour and variable sensitivity to targeted therapies (36). Lung cancer with an activating EGFR mutation is composed of heterogeneous tumor cell clones harbouring various combinations of EGFR gene alleles, including activating, wild type and resistant clones prior to treatment with EGFR-TKIs. Tumoral clonal composition is modified with EGFR-TKI treatments by clonal pressure and the appearance of new resistant clones (37).
Even in cases where one clone dominates, low frequency of subclones can determine clinical course and, depending on treatment sequence, affect therapy outcomes due to the presence of simultaneous drug-resistant clones and reexpansion of sensitizing clones (38). Although this phenomenon is not well understood and further investigation is needed.
In this situation, liquid biopsy reflects better tumor heterogeneity than single biopsies. Our center is being a pioneer in monitoring ctDNA in EGFR-lung cancer patients under EGFR-TKI treatment, obtaining ctDNA from plasma periodically and performing genotyping by digital PCR to detect more frequent alterations: exon 19 deletions and point mutations (L858R, G719X, and T790M resistance mutation). If T790M mutation is not detected at progression to first- and second-generation EGFR-TKI, or patients progress to Osimertinib, plasma NGS is performed to better understand mechanisms of resistance and offer the patient any therapeutic option if possible.
Conclusions
Here we reported a rare case simultaneously present with quadruple EGFR mutations: EGFR L858R/T790M/L718Q/C797S. Dynamic tumor mutation profiles and heterogeneity were monitored by performing periodic non-invasive liquid biopsy and ctDNA analyses, and a more comprehensive genetic profiling was acquired by the complementary NGS analyses on resection specimen. Based on these data, this case confirmed that EGFR L718Q independently led to osimertinib resistance, and an effective treatment strategy to this rare mutation was developed from the perspective of precision medicine.
Acknowledgments
Funding: Beijing Natural Science Foundation (7182132); Special Data Service for Oncology, The National Population and Health Scientific Data Sharing Platform (NCMI-ABD02-201809; NCMI-YF02N-201906), Supported by Ministry of Science and Technology of the People’s Republic of China (MOST); Chinese Academy of Medical Sciences Young Medical Talent Award Fund (no.2018RC320005); Beijing Students’ platform for innovation and entrepreneurship training program (2019zlgc0629); National Key Research and Development Program of China Grant (No. 2016YFC0901500).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2020.03.29). SZ reports grants from National Key Research and Development Program of China Grant, during the conduct of the study. NL reports grants from Beijing Natural Science Foundation, grants from Ministry of Science and Technology of the People’s Republic of China, grants from Chinese Academy of Medical Sciences Young Medical Talent Award Fund, grants from Beijing Students’ platform for innovation and entrepreneurship training program, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was obtained from the patient for publication of this case report and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu M, He WX, Song N, et al. Discrepancy of epidermal growth factor receptor mutation in lung adenocarcinoma presenting as multiple ground-glass opacities. Eur J Cardiothorac Surg 2016;50:909-13. [Crossref] [PubMed]
- Ma L, Chen R, Wang F, et al. EGFR L718Q mutation occurs without T790M mutation in a lung adenocarcinoma patient with acquired resistance to osimertinib. Ann Transl Med 2019;7:207. [Crossref] [PubMed]
- Callegari D, Ranaghan KE, Woods CJ, et al. L718Q mutant EGFR escapes covalent inhibition by stabilizing a non-reactive conformation of the lung cancer drug osimertinib. Chemical Science 2018.9-9. [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Bersanelli M, Minari R, Bordi P, et al. L718Q Mutation as New Mechanism of Acquired Resistance to AZD9291 in EGFR-Mutated NSCLC. J Thorac Oncol 2016;11:e121-3. [Crossref] [PubMed]
- Ercan D, Choi HG, Yun CH, et al. EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin Cancer Res 2015;21:3913-23. [Crossref] [PubMed]
- Knebel FH, Bettoni F, Shimada AK, et al. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer 2017;108:238-41. [Crossref] [PubMed]
- Zhang YC, Zhou Q, Wu YL. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J Hematol Oncol 2017;10:167. [Crossref] [PubMed]
- de Wit S, Rossi E, Weber S, et al. Single tube liquid biopsy for advanced non-small cell lung cancer. Int J Cancer 2019;144:3127-37. [Crossref] [PubMed]
- Madic J, Kiialainen A, Bidard FC, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer 2015;136:2158-65. [Crossref] [PubMed]
- Andree KC, Mentink A, Zeune LL, et al. Toward a real liquid biopsy in metastatic breast and prostate cancer: Diagnostic LeukApheresis increases CTC yields in a European prospective multicenter study (CTCTrap). Int J Cancer 2018;143:2584-91. [Crossref] [PubMed]
- Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med 2012;10:138. [Crossref] [PubMed]
- Stetson D, Ahmed A, Xu X, et al. Orthogonal Comparison of Four Plasma NGS Tests With Tumor Suggests Technical Factors are a Major Source of Assay Discordance. JCO Precision Oncology 2019;1-9.
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Siravegna G, Mussolin B, Venesio T, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol 2019;30:1580-90. [Crossref] [PubMed]
- Vendrell JA, Mau-Them FT, Béganton B, et al. Circulating Cell Free Tumor DNA Detection as a Routine Tool forLung Cancer Patient Management. Int J Mol Sci 2017;18:E264. [Crossref] [PubMed]
- Zugazagoitia J, Gómez-Rueda A, Jantus-Lewintre E, et al. Clinical utility of plasma-based digital next-generation sequencing in oncogene-driven non-small-cell lung cancer patients with tyrosine kinase inhibitor resistance. Lung Cancer 2019;134:72-8. [Crossref] [PubMed]
- Garcia-Pelaez B, Jantus E, Romero A, et al. TiPRING observational trial to compare T790M mutation testing in blood by different methodologies. Ann Oncol 1890;2018:29.
- Lindsay CR, Blackhall FH, Carmel A, et al. EPAC-lung: pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur J Cancer 2019;117:60-8. [Crossref] [PubMed]
- Pezzuto A, Manicone M, Scaini MC, et al. What information could the main actors of liquid biopsy provide? -a representative case of non-small cell lung cancer (NSCLC). J Thorac Dis 2018;10:E570-6. [Crossref] [PubMed]
- Cappuzzo F, Morabito A, Normanno N, et al. Efficacy and safety of rechallenge treatment with gefitinib in patients with advanced non-small cell lung cancer. Lung Cancer 99:31-7. [Crossref] [PubMed]
- Oda N, Hotta K, Ninomiya K, et al. A phase II trial of EGFR-TKI readministration with afatinib in advanced non-small-cell lung cancer harboring a sensitive non-T790M EGFR mutation: Okayama Lung Cancer Study Group trial 1403. Cancer Chemother Pharmacol 2018;82:1031-8. [Crossref] [PubMed]
- Ramalingam S, Gray J, Ohe Y, et al. LBA5_PROsimertinib vs comparator EGFR-TKI as first-line treatment for EGFRm advanced NSCLC (FLAURA): Final overall survival analysis. Ann Oncol 2019.30.
- Gray J, Peled N, Markovets A, et al. LBA85Longitudinal circulating tumour DNA (ctDNA) monitoring for early detection of disease progression and resistance in advanced NSCLC in FLAURA. Ann Oncol 2019.30.
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 21:3196-203. [Crossref] [PubMed]
- Metro G, Baglivo S, Siggillino A, et al. Successful Response to Osimertinib Rechallenge after Intervening Chemotherapy in an EGFR T790M-Positive Lung Cancer Patient. Clin Drug Investig 2018;38:983-7. [Crossref] [PubMed]
- Rossi E, Zamarchi R. Single-Cell Analysis of Circulating Tumor Cells: How Far Have We Come in the -Omics Era? Front Genet 2019;10:958. [Crossref] [PubMed]
- D'Antonio C, Milano A, Righini R, et al. Pharmacogenomics in lung cancer chemotherapy: a review of what the oncologist should know. 2014;34:5241-50.
- Danesi R, Altavilla G, Giovannetti E, et al. Pharmacogenomics of gemcitabine in non-small-cell lung cancer and other solid tumors. Pharmacogenomics 10:69-80. [Crossref] [PubMed]
- Tan LM, Qiu CF, Zhu T, et al. Genetic Polymorphisms and Platinum-based Chemotherapy Treatment Outcomes in Patients with Non-Small Cell Lung Cancer: A Genetic Epidemiology Study Based Meta-analysis. Sci Rep 2017;7:5593. [Crossref] [PubMed]
- Camps C, Sarries C, Roig B, et al. Assessment of Nucleotide Excision Repair XPD Polymorphisms in the Peripheral Blood of Gemcitabine/Cisplatin–Treated Advanced Non–Small-Cell Lung Cancer Patients. Clin Lung Cancer 2003;4:237-41. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 29:iv192-iv237. [Crossref]
- Nagano T, Tachihara M, Nishimura Y. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy. Cells 2018;7:212. [Crossref] [PubMed]
- Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clinical Cancer Research An Official Journal of the American Association for Cancer Research clincanres.2310.017.
- Wang S, Tsui ST, Liu C, et al. EGFR C797S mutation mediates resistance to third-generation inhibitors in T790M-positive non-small cell lung cancer. J Hematol Oncol 2016;9:59. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Imamura F, Uchida J, Kukita Y, et al. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 2016;94:68-73. [Crossref] [PubMed]
- Kohsaka S, Petronczki M, Solca F, et al. Tumor clonality and resistance mechanisms in EGFR mutation-positive non-small-cell lung cancer: implications for therapeutic sequencing. Future Oncol 2019;15:637-52. [Crossref] [PubMed]


