
WW and C2 domain-containing protein-3 promoted EBSS-induced apoptosis through inhibiting autophagy in non-small cell lung cancer cells
Introduction
Autophagy is considered to be an evolutionarily conservative process in all eukaryotic cells. It helps cells to maintain homeostasis, adapt to changing nutritional conditions, circulate cytoplasm, generate macromolecular building blocks and energy under pressure, and remove redundant and damaged proteins and organelles (1,2). Autophagy is believed to play a complex role in the occurrence, development, and treatment of lung cancer (3-5). For example, low Beclin-1 expression has been found to be significantly correlated with tumorigenesis, tumor progression, and poor prognosis in non-small cell lung carcinoma (NSCLC) (6). Rao et al. discovered that the development of NSCLC could be accelerated by inactivating autophagy-related 5 (ATG5), an important protein involved in autophagy (7). The inhibition of autophagy can weaken the proliferative ability of lung cancer cells and promote their sensitivity to chemotherapeutic drug-induced apoptosis (8). Although great progress has been made with our understanding of autophagy regulation to date, the detailed information about the regulation of autophagy remains limited.
WW and C2 domain-containing protein (WWC3) is a member of the WWC protein family (KIBRA/WWC1, WWC2, and WWC3), which maps to the human chromosomal locus Xp22.2 (9). Our previous studies demonstrated that low WWC3 expression is present in both lung cancer cell lines and lung cancer specimens and is associated with low differentiation, advanced pathological tumor-node-metastasis (pTNM) stage, positive lymph node metastasis, and poor prognosis in lung cancer patients. Meanwhile, the ectopic expression of WWC3 has an inhibitory role in the proliferation and invasiveness of lung cancer cells in vitro and in vivo (10,11). A recent study indicated that KIBRA/WWC1 is involved in autophagy processing in S2 cells and in Drosophila larvae (12). These results prompted us to explore the involvement of WWC3 in autophagy and apoptosis in lung cancer cells under starvation or hypoxic conditions.
In this study, we found that forced expression of WWC3 inhibited starvation-induced autophagy and promoted apoptosis of lung cancer cells. Our results provide valuable new insight into the mechanism by which the biological behavior of lung cancer is influenced by WWC3, which may serve as a potential target for the treatment of lung cancer patients.
Methods
Cell culture
The human bronchial epithelial (HBE) cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The NSCLC cell lines A549, H1299 and H460 were purchased from Shanghai Cell Bank (Shanghai, China). The LK2 cell line was a gift from Dr. Hiroshi Kijima (Department of Pathology and Bioscience, Hirosaki University Graduate School of Medicine, Japan). Upon receipt, the cells were frozen and individual aliquots were typically cultured for analysis within 10 passages. All cells were cultured in RPMI 1640 (Hyclone, Logan, UT, USA) containing 10% fetal calf serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 IU/mL penicillin, and 100 µg/mL streptomycin at 37 °C with 5% CO2 in high humidity. All cell lines were authenticated by short tandem repeat (STR) DNA profiling.
Plasmids, small interfering RNA (siRNA), and reagents
pEGFP-C2-WWC3 and the corresponding pEGFP-C2 empty vectors were provided by Dr. Joachim Kremerskothen (University of Münster, Germany). siRNA-WWC3 (sc-91139) and control siRNA (sc-37007) were purchased from Santa Cruz Technology Inc. (CA, USA). Lipofectamine 3000 (Thermo Fisher Scientific) transfection reagent was used for plasmid transfection. Earle’s balanced salt solution (EBSS, NaHCO3: 2.2 g/L, glucose: 1.0 g/L, phenol red: 0.011 g/L, #E2888), chloroquine (CQ, #C6628), and 3-methyladenine (3-MA, M9281) were all purchased from Sigma-Aldrich (St. Louis, MO, USA).
Western blot analysis
Total protein from the cell lines was extracted with lysis buffer (Thermo Fisher Scientific) and quantified using the Bradford method. SDS-PAGE (8% and 15%) was used to separate 40 µg of protein. The protein was transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) before incubation overnight at 4 °C with the following antibodies: WWC3 (#HPA039814, 1:1,000; Sigma-Aldrich); anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; sc-293335, 1:1,000; Santa Cruz Biotechnology); LC3B (#3868, 1:1,000); Beclin-1 (#3738, 1:1,000); P62 (#39749, 1:500); caspase-3 (#9662, 1:500); cleaved caspase-3 (#9664, 1:500); caspase-7 (#9494, 1:500); and cleaved caspase-7 (#9491, 1:500) (all from Cell Signaling Technology, Danvers, MA, USA). After incubation with peroxidase-coupled anti-mouse IgG and anti-rabbit IgG (Santa Cruz Biotechnology) at 37 °C for 2 h, bound proteins were viewed using enhanced chemiluminescence (ECL) (Thermo Fisher Scientific) and detected using BioImaging Systems (UVP Inc., Upland, CA, USA). The relative protein levels were calculated with GAPDH used as the loading control.
Quantitative real-time polymerase chain reaction (RT-qPCR) (SYBR Green method)
The assay was described in our previous study (10). The primer sequences used were: WWC3: forward, 5'-CAAGAGCGCATGTTGAAGGAA-3', reverse: 5'-CGCTGCTGCTTAATCTGGTAGA-3'; GAPDH: forward: 5'-GGAGCGAGATCCCTCCAAAAT-3', reverse: 5'-GGCTGTTGTCATACTTCTCATGG-3'.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, blocked with 3% BSA, and incubated with LC3B (#3868, 1:100, Cell Signaling Technology) overnight at 4 °C. Tetramethylrhodamine-5-(and-6)-isothiocyanate (TRITC)-conjugated secondary antibodies were then added for 1 h at room temperature. The cells were counterstained with 4',6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific). Epifluorescence microscopy was performed using an inverted Nikon TE300 microscope (Melville, NY, USA), and confocal microscopy was carried out with a Radiance 2000 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY, USA).
Labeling and capture of nascent RNA
Click-iT Nascent RNA Capture Kit (Thermo Fisher Scientific) was used to synthesize and isolate new RNA according to the manufacturer’s instructions. Briefly, cells were incubated in 0.2 mM of 5-ethynyl uridine (5-EU; an alkyne-modified uridine analog, which was efficiently and naturally incorporated into the nascent RNA) for 4 h, and the total RNA labeled with EU was isolated using TRIzol reagent (Invitrogen). Then, the EU-labeled RNA was biotinylated in a Click-iT reaction buffer with 0.5 mM of biotin azide and subsequently captured on streptavidin magnetic beads.
Detection of apoptosis
Apoptosis induced by EBSS and 3-MA was detected with the Annexin V-APC Apoptosis Detection Kit according to the manufacturer’s instructions (BD Biosciences Pharmingen, CA, USA). Briefly, 2×105 cells were washed once with ice-cold phosphate-buffered saline (PBS, pH 7.4) and resuspended in 200 mL of binding buffer. The cells were incubated with Annexin V-APC for 30 min and then with propidium iodide (PI) for 5 min at room temperature. Before testing, 400 mL of binding buffer was added. Apoptotic cells were detected by flow cytometry (FACS Calibur, Becton Dickinson, NJ, USA). All experiments were performed in triplicate.
Cell viability
Cells were plated in 96-well plates with medium containing 10% fetal bovine serum (FBS) (2,000 cells/well) 48 h after transfection. Cell viability was determined using the (3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTT) assay. Briefly, 20 µL of the 5 mg/mL MTT solution (Sigma, cell viability) was added to each well, and the mixture were incubated for 4 h at 37 °C. Then, the medium was removed from each well and the resultant MTT formazan was solubilized in 150 µL of dimethyl sulfoxide (DMSO). The results were repeated 5 times and quantified spectrophotometrically using a test wavelength of 490 nm.
Caspase-3/7 activity assay
Cell lysates were detected using 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer (50 mM HEPES, pH 7.4; 0.1% CHAPS, 0.1% Triton X-100, 0.1 mM EDTA, and 1 mM dithiothreitol) and centrifuged at 12,000 g at 4 °C. DEVDase activity was measured using the specific fluorogenic substrate, Asp-Glu-Val-Asp-7-amino-4-methylcoumarin (DEVD-AMC, Sigma-Aldrich) in the assay buffer (50 mM HEPES, pH 7.4; 100 mM NaCl, 0.1% CHAPS, 10% glycerol, 1 mM EDTA, and 10 mM dithiothreitol), and a fluorescence plate reader was used to obtain real-time recordings of enzyme-catalyzed AMC release. Fluorescence values were converted into picomoles of released AMC using the standard curve generated from free AMC, and the maximum rate of AMC release (pmol/min/mg protein) was calculated. All experiments were performed in triplicate.
Statistical analysis
SPSS 22.0 (SPSS, Chicago, IL, USA) was used to perform all the statistical analyses. The statistical significance of the differences was analyzed by Student’s t-test. All experiments were performed in triplicate, and the results were presented as the mean ± standard deviation (SD). A P value <0.05 indicated statistical significance.
Results
Starvation could inhibit WWC3 expression in NSCLC cells
To explore the role of WWC3 in autophagy in NSCLC cells, the expression level of WWC3 in A549 and H460 cell lines cultured with complete EBSS, the most direct inducer of typical autophagy, was detected at different time points (0, 6, 12 and 24 h). In A549 and H460 cells, we observed that the WWC3 protein level had obviously decreased after 6 h of culture with complete EBSS medium. The lowest expression level in the A549 and H460 cells was reached at 12 and 24 h, respectively (Figure 1A,B). There was also a marked decrease in WWC3 mRNA expression after culture with complete EBSS (Figure 1C,D). Because the decrease of WWC3 mRNA level after complete EBSS culture may be attributed to the decrease of mRNA synthesis, a new WWC3 transcription gene was isolated from the A549 and H460 cells using Click-iT Nascent RNA Capture, and analyzed. The results of RT-qPCR showed that starvation-induced by complete EBSS could significantly reduce WWC3 mRNA expression (Figure 1E). Also, when combined with 20 µM CQ, the lysosomal conversion inhibitor did not alter the reduction of WWC3 in cells cultured with complete EBSS (Figure 1F), which suggests that the activation of autophagy degradation may not promote the reduction of WWC3 mediated by EBSS starvation in lung cancer cells.
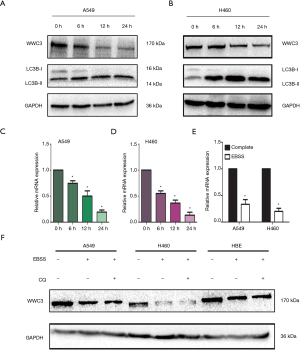
WWC3 attenuated EBSS starvation-induced autophagy in NSCLC
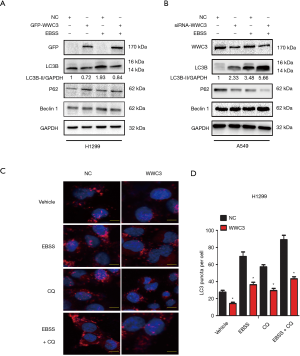
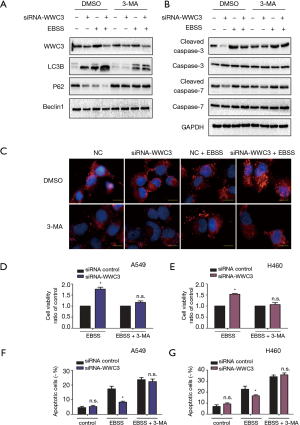
The downregulation of WWC3 caused by EBSS starvation led us to explore the potential role of WWC3 involved in starvation-induced typical autophagy. H1299 cells, which have been reported to have low WWC3 expression (10), were transfected with GFP-WWC3 plasmid and then cultured with normal medium or in complete EBSS media for 14 h. The immunoblotting assay demonstrated that WWC3 overexpression eliminated the increased basal LC3-II expression induced by starvation. Under the same conditions, P62/SQSTM1 expression was enhanced in the H1299 cell line and, notably, Beclin1 expression level was not noticeably altered (Figure 2A). In contrast, knockdown of WWC3 by siRNA-WWC3 in A549 cells, a cell line known to have a relatively high expression of WWC3, exerted the opposite effect (Figure 2B). The obvious punctate distribution of endogenous LC3, a marker for autophagosome formation, was also investigated. The results of the immunofluorescence assay showed that forced WWC3 expression was markedly different from the basal punctuate distribution in H1299 cells. Moreover, when WWC3 transfected cells were cultured in complete EBSS, even in the presence of 20 µM CQ for 12 h, the number of LC3 puncta per cell was significantly reduced (Figure 2C,D). These results indicate that WWC3 has a Beclin1-independent inhibitory effect on starvation-induced autophagy in lung cancer cells.
WWC3 promoted EBSS-induced cell apoptosis in NSCLC
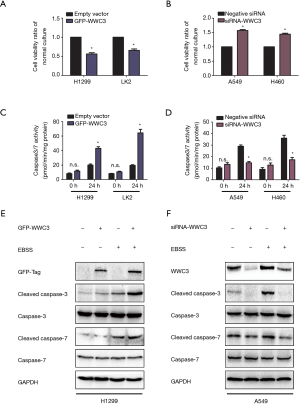
To explore the potential effect of WWC3 on lung cancer cell growth under starvation conditions, we performed an MTT proliferation assay. Compared with the empty vector group, ectopic WWC3 expression substantially inhibited the proliferation of H1299 and LK2 cells when cultured in complete EBSS media, whereas the proliferative ability of A549 and H460 cells was significantly enhanced by WWC3 silencing under starvation conditions (Figure 3A,B). The impact of WWC3 on the activity of caspase-3/7 was also investigated. WWC3 transfection significantly enhanced caspase-3/7 activity under starvation conditions in H1299 and LK2 cells, while WWC3 knockdown in A549 and H460 cells had the opposite effect (Figure 3C,D). The immunoblotting assay demonstrated that WWC3 increased the expression of apoptosis-related molecules, cleaved caspase-3 and cleaved caspase-7 (Figure 3E,F); these results were consistent with those of the caspase-3/7 activity assay. Additionally, we adopted the Annexin-V APC/PI staining method to detect the impact of ectopic WWC3 overexpression on EBSS-induced apoptosis of lung cancer cells. The results showed that WWC3 could promote EBSS-induced apoptosis of H1299 cells (Figure S1). Therefore, WWC3 could potentially be involved in the apoptosis regulation process under EBSS starvation conditions.
The impact of WWC3 knockdown on cell apoptosis under starvation was attenuated by autophagy inhibition
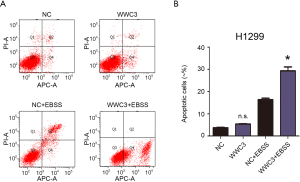
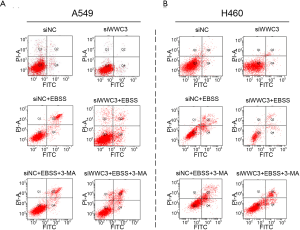
Previous studies have shown that autophagy activation caused by starvation or hypoxia is a mechanism for promoting survival, thus inhibiting cell death (13). Therefore, we hypothesized that WWC3 could promote starvation-induced apoptosis by attenuating autophagy. The effect 3-MA, which is considered to be a negative regulator of PtdIns3K and is widely used as an inhibitor of hunger-mediated autophagy at a preliminary stage, was determined (14,15). We transfected siRNA-WWC3 into A549 cells to silence the WWC3 expression. Western blotting showed that under the media with 3-MA (5 µM) for 24 h, starvation-mediated P62/SQSTM1 and LC3-II transition expression was inhibited in both the control and WWC3-silenced A549 cells (Figure 4A). We also investigated the impact of 3-MA on apoptosis-related molecules (cysteinyl aspartate-specific proteinases, caspase-3 and caspase-7) after WWC3 knockdown under complete EBSS culture conditions. We found that cleaved caspase-3/7 were significantly downregulated after transfection with siRNA-WWC3 under normal or starvation conditions and that the presence of 3-MA reversed this tendency (Figure 4B). The immunofluorescence assay revealed that treatment with 5 µM 3-MA for 24 h also reversed the increased LC3 punctate distribution induced by starvation in both the control and knockdown groups (Figure 4C). Moreover, compared with control group, the inhibition of autophagy caused by 3-MA significantly reversed the increase in cell viability under starvation conditions in A549 and H460 WWC3-silenced cells (Figure 4D,E). fluorescence-activated cell sorting (FACS) results showed that treatment with 5 µM 3-MA for 24 h restored the decreased apoptotic rate of A549 and H460 lung cancer cells induced by WWC3 knockdown (Figures 4F,G,S2). Taking the above results into account, we believe that WWC3 promotes the apoptosis of lung cancer cells induced by starvation, at least partially through the attenuation of autophagy.
Discussion
Autophagy and apoptosis are important physiological processes in cells, and are vital for cell survival and growth (16,17). Autophagy is the key process by which tumorigenesis, tumor stromal interaction, and the effect of tumor therapy are regulated, but it plays a dual and environment-dependent role in tumor inhibition and survival (18). An increasing amount of research has shown that, in some cases, autophagy can protect malignant cells (19-22), while in other cases, autophagy is related to type II programmed cell death (23,24). Understanding and elucidating the molecular mechanism of autophagy, as well as how autophagy can be regulated to kill tumor cells, are still important focuses of tumor therapy. Hence, clarifying the role of WWC3 in autophagy, the role autophagy plays in cell viability, and the mechanisms of autophagy regulation is vital for the development of WWC3-targeted drugs for NSCLC treatment.
The three members of the WWC protein family, KIBRA/WWC1, WWC2, and WWC3, are the upstream molecules of the Hippo signaling pathway (9). Our research and other scholars have shown that WWC3 has an anti-proliferation effect on solid tumor cell lines such as lung cancer, glioma, and gastric cancer (10,11,25-27). To date, only Jin et al. have reported the crucial roles that KIBRA/WWC1 and aPKC perform in regulating starvation-induced autophagy (12), and studies addressing the relationship between WWCs and autophagy are limited. In the present study, we demonstrated that WWC3 served as a negative regulator of autophagy under starvation-induced stress in lung cancer cells, and promoted their apoptosis rate.
First, we found that the expression of WWC3, including at the mRNA and protein levels, markedly decreased in lung cancer cells when cultured in EBSS media. Since the activation of autophagy can promote protein degradation, we then added autophagy inhibitor 3-MA to starvation-conditioned cells. We found that the activation of autophagic degradation of lung cancer cells may not be related to the EBSS starvation-mediated reduction of WWC3. This may be attributed to the inadequate energy intake of lung cancer cells caused by starvation and the obstacle of raw material synthesis.
Subsequently, we examined the effect of WWC3 on the expression of autophagy-related biomarkers, LC3-II and P62, under starvation. The results showed that overexpression of WWC3 could significantly downregulate the expression of LC3-II and inhibit the degradation of the P62 protein. Immunofluorescence also demonstrated that WWC3 overexpression could decrease the number of endogenous LC3 fluorescent spots, which was consistent with the results of the Western blotting. Remarkably, the expression of autophagy-related protein Beclin-1 did not change significantly during this process. This suggests that WWC3 influences autophagy via Beclin-1-independent pathways. However, the specific molecular mechanisms on which WWC3 relies remain unclear.
The WWC protein family members are known to act as classical upstream proteins of the Hippo signaling pathway; they suppress yes-associated protein (YAP) transcriptional activity and trigger Hippo pathway activity. A number of studies have reported the relationship between the Hippo pathway and autophagy (28-30). Whether WWC3 can inhibit autophagy via the Hippo pathway is a question that will be answered in future research.
Finally, we performed FACS assay and Western blotting to demonstrate that WWC3 knockdown could decrease the expression of apoptosis-related molecules such as active caspase-3/7, and hence inhibit the apoptotic rate of starved lung cancer cells. The presence of 3-MA abrogated this effect, indicating that WWC3 enhanced apoptosis of NSCLC cells caused by EBSS starvation through inhibiting autophagy. It is worth noting that we used siRNA to knock down WWC3 expression to observe the effect of decreased expression of WWC3 on the EBSS-induced apoptosis rate and the apoptosis-related protein expression in A549 cell line. The results showed that EBSS-induced apoptosis could be attenuated by silencing WWC3. Furthermore, we also observed that EBSS could increase the cleaved caspase-3/7 expression in DMSO-treated WWC3 knockdown cells, which may be related to the action time of EBSS. The best time to detect protein changes in the signal pathway with Western blotting is quite different from that for apoptosis assay. In general, the change of protein precedes the appearance of cell phenotype. Therefore, we believe that while the apoptosis-related protein caspase-3/7 can be dramatically upregulated in lung cancer cells after EBSS treatment for 24 h, the degree of apoptosis may not be significantly changed. We also calculated the statistical difference in DMSO-treated WWC3 knockdown cells after EBSS treatment in Figure 4F, and in fact, the results showed statistical significance. Combined with the results of Figure 3E,F, caspase-3/7 may be closely related to EBSS-induced apoptosis. Undoubtedly, adding caspase-3/7 inhibitor is the most direct way to prove this view. We will attempt to explain and verify this in our future research. To conclude, under tumor cell starvation, by upregulating some mechanisms of WWC3 and combining with 3-MA drugs, it is expected to become a new targeted treatment strategy for lung cancer.
Conclusions
In summary, we demonstrated that WWC3 promoted EBSS starvation-induced apoptosis of NSCLC cells via inhibiting autophagy. 3-MA can potentially be applied in lung cancer treatment strategies to sensitize cells and resist WWC3-targeted therapy by blocking the downregulation of WWC3-mediated protective autophagy.
Acknowledgments
We thank Professor Joachim Kremerskothen for kindly providing the pEGFP-C2 and pEGFP-C2-WWC3 plasmids.
Funding: This study was supported by the National Natural Science Foundation of China (grant no. 81572854 and 81772489 to EW; grant no. 81902986 to QH) and the China Postdoctoral Science Foundation (grant no. 2018M641737 to QH).
Footnote
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-966
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-966). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Su Z, Wang K, Li R, et al. Overexpression of RBM5 induces autophagy in human lung adenocarcinoma cells. World J Surg Oncol 2016;14:57. [Crossref] [PubMed]
- Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res 2014;24:24-41. [Crossref] [PubMed]
- Meng J, Chang C, Chen Y, et al. EGCG overcomes gefitinib resistance by inhibiting autophagy and augmenting cell death through targeting ERK phosphorylation in NSCLC. Onco Targets Ther 2019;12:6033-43. [Crossref] [PubMed]
- Fan J, Zhang X, Wang S, et al. Regulating autophagy facilitated therapeutic efficacy of the sonic Hedgehog pathway inhibition on lung adenocarcinoma through GLI2 suppression and ROS production. Cell Death Dis 2019;10:626. [Crossref] [PubMed]
- Wang K, Chen B, Yin T, et al. N-methylparoxetine blocked autophagic flux and induced apoptosis by activating ROS-MAPK pathway in non-small cell lung cancer cells. Int J Mol Sci 2019;20:3415. [Crossref] [PubMed]
- Zhou W, Yue C, Deng J, et al. Autophagic protein Beclin1 serves as an independent positive prognostic biomarker for non-small cell lung cancer. PLoS One 2013;8:e80338. [Crossref] [PubMed]
- Rao S, Yang H, Penninger JM, et al. Autophagy in non-small cell lung carcinogenesis: a positive regulator of antitumor immunosurveillance. Autophagy 2014;10:529-31. [Crossref] [PubMed]
- Kaminskyy VO, Piskunova T, Zborovskaya IB, et al. Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy 2012;8:1032-44. [Crossref] [PubMed]
- Wennmann DO, Schmitz J, Wehr MC, et al. Evolutionary and molecular facts link the WWC protein family to Hippo signaling. Mol Biol Evol 2014;31:1710-23. [Crossref] [PubMed]
- Han Q, Lin X, Zhang X, et al. WWC3 regulates the Wnt and Hippo pathways via Dishevelled proteins and large tumour suppressor 1, to suppress lung cancer invasion and metastasis. J Pathol 2017;242:435-47. [Crossref] [PubMed]
- Han Q, Kremerskothen J, Lin X, et al. WWC3 inhibits epithelial-mesenchymal transition of lung cancer by activating Hippo-YAP signaling. Onco Targets Ther 2018;11:2581-91. [Crossref] [PubMed]
- Jin A, Neufeld TP, Choe J. Kibra and aPKC regulate starvation-induced autophagy in Drosophila. Biochem Biophys Res Commun 2015;468:1-7. [Crossref] [PubMed]
- Yogev O, Goldberg R, Anzi S, et al. Jun proteins are starvation-regulated inhibitors of autophagy. Cancer Res 2010;70:2318-27. [Crossref] [PubMed]
- Li S, Zhang HY, Wang T, et al. BAG3 promoted starvation-induced apoptosis of thyroid cancer cells via attenuation of autophagy. J Clin Endocrinol Metab 2014;99:E2298-307. [Crossref] [PubMed]
- Grishchuk Y, Ginet V, Truttmann AC, et al. Beclin-1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy 2011;7:1115-31. [Crossref] [PubMed]
- Manu KA, Cao PHA, Chai TF, et al. p21cip1/waf1 coordinate autophagy, proliferation and apoptosis in response to metabolic stress. Cancers 2019;11:1112. [Crossref] [PubMed]
- Emdad L, Bhoopathi P, Talukdar S, et al. Recent insights into apoptosis and toxic autophagy: The roles of MDA-7/IL-24, a multidimensional anti-cancer therapeutic. Semin Cancer Biol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Weckman A, Rotondo F, Di Ieva A, et al. Autophagy in endocrine tumors. Endocr Relat Cancer 2015;22:R205-18. [Crossref] [PubMed]
- Rabinowitz JD, White E. Autophagy and metabolism. Science 2010;330:1344-8. [Crossref] [PubMed]
- Jin P, Jiang J, Xie N, et al. MCT1 relieves osimertinib-induced CRC suppression by promoting autophagy through the LKB1/AMPK signaling. Cell Death Dis 2019;10:615. [Crossref] [PubMed]
- Banach A, Jiang YP, Roth E, et al. CEMIP upregulates BiP to promote breast cancer cell survival in hypoxia. Oncotarget 2019;10:4307-20. [Crossref] [PubMed]
- Kim MJ, Hwang GY, Cho MJ, et al. Depletion of NBR1 in urothelial carcinoma cells enhances rapamycin-induced apoptosis through impaired autophagy and mitochondrial dysfunction. J Cell Biochem 2019;120:19186-201. [Crossref] [PubMed]
- Li F, Guo H, Yang Y, et al. Autophagy modulation in bladder cancer development and treatment Oncol Rep 2019;42:1647-55. (Review). [PubMed]
- Strappazzon F, Di Rita A, Peschiaroli A, et al. HUWE1 controls MCL1 stability to unleash AMBRA1-induced mitophagy. Cell Death Differ 2020;27:1155-68. [Crossref] [PubMed]
- Wang Y, Jiang M, Yao Y, et al. WWC3 inhibits glioma cell proliferation through suppressing the Wnt/β-catenin signaling pathway. DNA Cell Biol 2018;37:31-7. [Crossref] [PubMed]
- Ou C, Li X, Li G, et al. WWC3: the bridge linking Hippo and Wnt pathways in lung cancer. J Thorac Dis 2017;9:2315-6. [Crossref] [PubMed]
- Hou J, Zhou J. WWC3 downregulation correlates with poor prognosis and inhibition of Hippo signaling in human gastric cancer. Onco Targets Ther 2017;10:2931-42. [Crossref] [PubMed]
- Zhou Y, Wang Y, Zhou W, et al. YAP promotes multi-drug resistance and inhibits autophagy-related cell death in hepatocellular carcinoma via the RAC1-ROS-mTOR pathway. Cancer Cell Int 2019;19:179. [Crossref] [PubMed]
- Wang P, Gong Y, Guo T, et al. Activation of Aurora A kinase increases YAP stability via blockage of autophagy. Cell Death Dis 2019;10:432. [Crossref] [PubMed]
- Pei T, Huang X, Long Y, et al. Increased expression of YAP is associated with decreased cell autophagy in the eutopic endometrial stromal cells of endometriosis. Mol Cell Endocrinol 2019;491:110432. [Crossref] [PubMed]


