
Identification of key immune-related genes and immune infiltration in atrial fibrillation with valvular heart disease based on bioinformatics analysis
Introduction
Atrial fibrillation (AF) is the most common persistent arrhythmia (1). Patients who develop AF have an increased risk of heart failure, stroke, cardioembolic hospitalization, and mortality (1). Patients with valvular heart disease (VHD) frequently develop AF, because VHD can lead to the disruption of the atrial architecture due to increased left atrial (LA) pressure. It can also cause atrial dilation and alter wall stress (2). VHD patients, especially those with rheumatic mitral stenosis, have a significant risk of stroke and thromboembolism. On average, the rate of stroke among patients with AF is approximately 6 times that of individuals without AF. The relative risk is even higher in patients with both AF and mitral stenosis (about 15 times that of people without AF). However, the underlying mechanisms of AF-VHD are not well understood. Therefore, understanding the AF-VHD biology is vital for the diagnosis and treatment of this complicated disease.
Previous studies have shown that immune and inflammatory responses may be essential in the pathophysiological mechanism of AF, also in AF-VHD, one of the main types of AF (3,4). Several studies have found that inflammatory biomarkers, such as C-reactive protein, interleukin (IL)-6, IL-10, and tumor necrosis factor (TNF)-α, have been found to be related to AF and its outcome (5-8). And some researches indicated that the elevated levels of serum hs-CRP, IL-6, and sCD-40L were strongly associated with AF-VHD (4,9,10). In addition, several immune-related genes (IRGs), such as C-X-C motif chemokine receptor (CXCR) 2, may play an important functional role in atrial remodeling and AF inducibility. The results of the above studies suggest that IRGs may be a new therapeutic strategy for AF (11,12). The infiltration of immune cells has also been demonstrated to influence the development of AF. Hohmann et al. reported the significant increase of inflammatory CD3+ T-cell counts over the course of sinus rhythm (SR) development into paroxysmal AF and persistent AF in LA appendages (LAA) (13). Yamashita et al. found that immune cells mostly consisted of immunologically active monocytes/macrophages, with CD3+ T cells playing a smaller role in human LAA with AF (14). Based on the potential the relationship between inflammation and AF, several studies have demonstrated that anti-inflammatory therapies, such as statins, steroids, or vitamin C, can significantly reduce AF episodes (15-17).
In the present study, we performed a bioinformatics analysis of IRGs in AF-VHD and investigated the underlying function and potential regulatory mechanisms of these IRGs. We used CIBERSORT to study the differential immune infiltration between 22 immune cell subsets in LA tissues from AF-VHD and SR patients (18). Furthermore, to better understand the underlying molecular immune mechanism of AF-VHD, we investigated the relationship between key IRGs and infiltrating immune cells. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-168).
Methods
Patients and datasets
The GSE41177 and GSE79768 datasets were downloaded from the Gene Expression Omnibus (GEO) database (GPL570 platform, HG-U133_Plus_2, Affymetrix Human Genome U133 Plus 2.0 Array). Only human LA samples from AF-VHD patients and VHD patients with SR (as the control group) were selected in each dataset. The GSE41177 contained 23 samples, including 16 LA tissue samples from patients with chronic AF (AF-VHD group) and 3 from patients with SR. The GSE79768 contained 7 atrial tissue samples from AF-VHD patients and 6 atrial tissue samples from patients with SR.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identifying differentially expressed IRGs (DEIRGs)
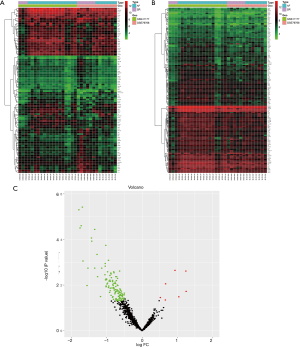
Gene expression matrices were created with R software (version 3.6.3; https://www.r-project.org/), and the R package of “Bioconductor” (http://www.bioconductor.org/) was applied for data analyses. The SVA package (https://www.bioconductor.org/packages/release/bioc/html/sva.html) was used for the normalization of the GSE41177 and GSE79768 datasets (19). Using the limma package (https://www.bioconductor.org/packages/release/bioc/html/limma.html), differentially expressed genes (DEGs) were screened. Only genes that satisfied the cut-off value of P<0.05 and fold change (FC) ≥1.5 (|log2 FC| >0.5) were selected as DEGs. DEIRGs were identified by matching 1,793 IRGs to DEGs using the IRG dataset from the ImmPort database (https://www.immport.org/shared/genelists). With the “pheatmap” package in R software (https://bioconductor.org/packages/release/bioc/html/heatmaps.html), heatmaps for the DEGs and DEIRGs was created. Using the “ggplot2” package, a volcano map of DEIRGs was created to show the differential expression of DEIRGs.
Functional and pathway enrichment analyses of DEIRGs
Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of DEIRGs was performed by using Database for Annotation, Visualization, and Integrated Discovery (https://david.ncifcrf.gov/) (20,21). P<0.05 illustrated statistical significance.
Protein-protein interaction (PPI) network and potential crucial genes analyses
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) plugin was applied to investigate the potential interaction between DEIRGs (confidence score: 0.9, maximum number of interactions: 0) and visualized the PPI network using Cytoscape software (version 3.8.1; https://www.cytoscape.org/) (22).
Evaluation of immune cell infiltration
Using CIBERSORT in R, we filtered out samples with P<0.05 and got an immune cell infiltration matrix with 22 types of immune cells. The proportions of immune cells were also obtained. The “ggplot2” package was conducted to carried out principal component analysis (PCA) and clustering analysis on the immune cell infiltration matrix data and created a 2D PCA map and violin diagrams to visualize the differences in immune cell infiltration. Spearman correlation analysis in R was applied to the 22 types of infiltrating immune cells, and the “Corrplot” package was conducted to create a correlation heatmap to visualize the result (23). The immune infiltration levels for each immune cell in the 2 groups were represented with violin diagrams using “the “vioplot” package in R.
Correlation analysis between key IRGs and infiltrating immune cells
The Spearman correlation analysis in R was used to explore the potential relations between the key IRGs and infiltrating immune cells, which was visualized by applying the package of “ggpubr”.
Results
DEIRG screening
After batch correction and standardization of the merged GSE41177 and GSE79768 datasets, a total of 919 DEGs, comprising 88 upregulated and 831 downregulated genes, were obtained (Figure 1A). Next, by matching the IRGs to the DEGs, 130 DEIRGs were selected, including 7 upregulated and 123 downregulated genes (Figure 1B,C).
GO analysis and pathway enrichment
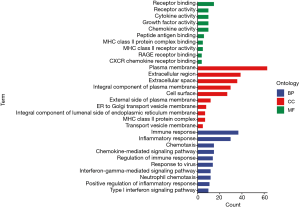
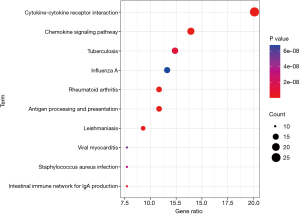
The results of the GO analysis showed that DEIRGs were mostly related to immune and inflammatory response under biological processes (BP), the plasma membrane and extracellular region under cellular components (CC), and receptor binding and receptor activity under molecular functions (MF) (Figure 2). The KEGG analysis results suggested that the DEIRGs were mostly enriched in cytokine-cytokine receptor interactions, antigen processing and presentation, rheumatoid arthritis (RA), the chemokine signaling pathway, and Leishmaniasis (Figure 3).
PPI network analysis
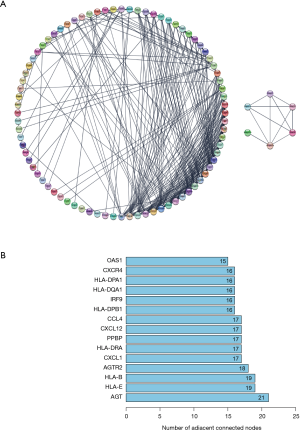
The PPI network of DEIRGs is shown in Figure 4. Degree (number of adjacent connected nodes) ≥15 was set as the cut-off criterion for hub genes (Figure 4B). Angiotensinogen (AGT), major histocompatibility complex class IE (HLA-E), HLA class IB (HLA-B), angiotensin II receptor type 2 (AGTR2), C-X-C motif chemokine ligand (CXCL) 1, HLA class II DR alpha (HLA-DRA), pro-platelet basic protein (PPBP), CXCL12, C-C motif chemokine ligand 4 (CCL4), HLA class II DP beta 1 (HLA-DPB1), interferon regulatory factor 9 (IRF9), HLA class II DQ alpha 1 (HLA-DQA1), HLA class II DP alpha 1 (HLA-DPA1), CXCR4, and 2'-5'-oligoadenylate synthetase 1 (OAS1) were identified as hub genes. The hub genes involved in both the immune response and inflammatory response in BP were selected as key genes, specifically CXCL1, PPBP, CXCL12, and CCL4.
Immune infiltration analyses
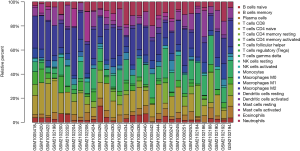
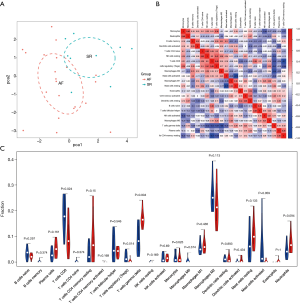
We first investigated the difference between AF-VHD and SR LA tissues in immune infiltration with 22 subpopulations of immune cells by using the CIBERSORT algorithm. The proportions of immune cells obtained from the 9 SR patients and 22 AF-VHD patients are summarized in Figure 5. Using PCA, the immune cells in the LA tissues of AF-VHD patients and SR revealed distinct group-bias clustering and individual differences (Figure 6A). The correlation heatmap showed that resting CD4 memory T cells and CD8 T cells, M2 macrophages, gamma delta T cells, activated mast cells, and resting mast cells all had a significant negative correlation. In contrast, naive CD4 T cells and resting natural killer cells had a significant positive correlation (correlation coefficients ≥0.5 or ≤–0.5 with P<0.05 were supposed to be the statistically significant positive or negative correlations) (Figure 6B). Compared with SR, LA tissues from AF-VHD patients generally contained a lower proportion of CD8 T cells and regulatory T cells (Tregs), whereas the proportions of gamma delta T cells were relatively higher (P<0.05) (Figure 6C).
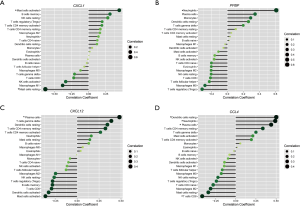
Correlation analysis between hub genes and infiltrating immune cells
The results of the correlation analysis revealed that CXCL1 was positively correlated with activated mast cells (r=0.441, P=0.013) and significantly negatively correlated with resting mast cells (r=–0.638, P=0.0001). PPBP was correlated significantly positively with neutrophils (r=0.603, P=0.0004). CXCL12 was positively correlated with plasma cells (r=0.476, P=0.007). CCL4 was positively correlated with plasma cells (r=0.432, P=0.016), neutrophils (r=0.483, P=0.007), and resting dendritic cells (r=0.494, P=0.005), and correlated negatively with CD8 T cells (r=0.483, P=0.019) (Figure 7).
Discussion
AF is one of the most prevalent arrhythmias (24). VHD and AF frequently coexist. When associated with VHD, the incidence of thromboembolism in AF significantly increases (25). Heart failure and stroke caused by thromboembolism are complications of AF that result in a high rate of disability and mortality, making AF a big challenge for public health (26). Treatments for AF in symptomatic patients include oral anticoagulants and LAA closure for stroke prevention, drugs for rate control, and anti-arrhythmic drug therapy and catheter ablation for conversion and the prevention of recurrence (2,27). However, the long-term efficacy and safety of these treatment regimens are still not well understood. The pathogenesis and biomarkers of AF still require further investigation. With better understanding of the underlying mechanisms of AF and AF-VHD, AF-VHD patients will receive better management and therapy. In recent years, increasing evidence has revealed that the pathogenesis of AF may be related to immune and inflammatory responses (28-31). Therefore, we sought to identify IRGs and explore the role of immune cell infiltration in AF-VHD in depth. A total of 130 DEIRGs were detected as candidate biomarkers in the pathogenesis of AF-VHD, and the enrichment function identified the underlying mechanisms of DEIRGs. The results of the GO enrichment analysis indicated that DEIRGs were significantly correlated with immune and inflammatory responses. Further, these genes were involved in cytokine-cytokine receptor interactions, the chemokine signaling pathway, tuberculosis, influenza A, RA, and other pathways via the KEGG analysis. Higher levels of cytokines, such as IL-6, IL-8, and TNF-α, have been reported in patients with AF compared with the levels in SR patients, and these inflammatory markers can predict the outcome of AF ablation (8,32-36). Similar with AF, the expression of some cytokines, such as IL-1β, TNF-α, TGF-β1 have increased in AF-VHD (37,38). Among them, the increase of TGFβ1 can promote the occurrence of atrial fibrosis, which is a key role in the mechanism of the occurrence and maintenance in atrial fibrillation (38-41). Recently, a Danish cohort study found that, compared with the risk among the general population, there is a 40% increase in the risk of AF in patients with RA (42). These findings indicate that DEIRGs in AF-VHD are involved in the immune and inflammatory processes for the disease.
The PPI was further analyzed by the STRING plugin in Cytoscape 3.8.1, and 15 hub genes (AGT, HLA-E, HLA-B, AGTR2, CXCL1, HLA-DRA, PPBP, CXCL12, CCL4, HLA-DPB1, IRF9, HLA-DQA1, HLA-DPA1, CXCR4, and OAS1) were selected in the present study. Of these, CXCL1, PPBP, CXCL12, and CCL4 were judged to be the core genes for the immune and inflammatory responses, according to the results of GO analysis.
CXCL1, a member of the CXC subfamily of chemokines, is a secreted growth factor that signals through the G-protein coupled receptor CXCR2 and modulates inflammation, angiogenesis, wound healing, tumorigenesis, and cell motility by regulating CXCL1–CXCL2 (43). Recent studies have indicated that CXCL1–CXCR2 signaling may play a crucial role in regulating monocyte infiltration and in the pathophysiology of hypertension and cardiac remodeling (44-46). It was also reported that the CXCL1/2 and CXCL1-CXCL2 single axis may participate in AF induced by a high dose of angiotensin II or prolonged hypertension (47). PPBP (also known as CXCL7), a platelet-derived growth factor, belongs to the CXC chemokine family. It is released from the activated platelets and participates in the response to vascular injury (48). It can also stimulate a variety of processes, such as mitogenesis, glucose metabolism, and the synthesis of the extracellular matrix and plasminogen activator (49,50). Maneerat et al. identified PPBP as one of the significant biomarkers correlated with the development of coronary heart disease (CHD) (51).
CXCL12 (also known as SDF-1) belongs to the CXC family, a subclass of the chemokine family, and regulates cellular activity by binding to CXCR4 or CXCR7, which are receptor proteins located in the cytomembrane, contributing to the activation of various intracellular signaling pathways (52-56). Previously published studies have suggested that there may be associations between CXCL12 and cardiovascular disease, leading to the elevation of serum CXCL12 levels, which in turn increases the risk of coronary artery disease (57-59). CCL4 (also known as macrophage inflammatory protein (MIP)-1beta, MIP-1β) is a crucial chemotactic mediator for the recruitment of monocytes/macrophages (60). It is associated with the pathogenesis of several diseases, including psoriasis vulgaris, sarcoidosis, cystic fibrosis, and multiple sclerosis (61-64). In obese patients, as well as in patients with a history of cardiac surgery, CCL4 levels have been found to be increased, which may be associated with new-onset AF (65,66). Moreover, it is also considered to play an important role in metabolic inflammation (67). Further research is required to explore the involvement of these 4 key IRGs in the mechanism of AF.
To further explore the role of immune cell infiltration in AF-VHD, CIBERSORT was used to evaluate AF-VHD immune infiltration. In this program, we found reduced infiltration of CD8 T cells and Tregs, as well as increased infiltration of gamma delta T cells, which may be related to the pathogenesis of AF-VHD. Several studies have shown that only a few CD8+ T cells are present in the left appendages of AF patients (14) . Chen et al. reported that, in peripheral whole blood and right atrial tissue of AF patients, the proportion of Treg cells was significantly lower (68). Nevertheless, to the best of our knowledge, there has been no research on the role of gamma delta T cells in AF-VHD and further experimental data are needed.
We further investigated the association between CXCL1, PPBP, CXCL12, CCL4, and immune infiltration. We found that CCL4 was positively correlated with plasma cells, neutrophils, and resting dendritic cells, and negatively correlated with CD8 T cells. CXCL1 was positively correlated with activated mast cells and significantly negatively correlated with resting mast cells. CXCL12 was positively correlated with plasma cells, and PPBP was significantly positively correlated with neutrophils. These findings require further researches to elucidate the underlying complicated interactions between IRGs and immune cells. Based on these results, CXCL1, PPBP, CXCL12, and CCL4 appear to play key roles in AF-VHD via the regulation of immune infiltration.
Conclusions
In summary, we found that the IRGs CXCL1, PPBP, CXCL12, and CCL4 may be key biomarkers of AF-VHD. We also found that some immune cells may contribute to the occurrence and progression of AF-VHD. Moreover, the correlations between CXCL1, PPBP, CXCL12, CCL4, and immune cells may play an important role in the pathogenesis of AF-VHD. Further researches of these IRGs and immune cells are required to study if they may be a novel agent to the molecular targeted therapies for AF-VHD patients.
Acknowledgments
We acknowledge GEO database for providing their platforms and contributors for uploading their meaningful datasets. And we thank all participants involved in this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-168
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-168). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). GEO belongs to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for researches and publish relevant articles. Our study is based on this open source data, so there are no ethical issues and other conflicts of interest.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Benjamin EJ, Levy D, Vaziri SM, et al. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. JAMA 1994;271:840-4. [Crossref] [PubMed]
- January CT, Wann LS, Alpert JS, et al. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. J Am Coll Cardiol 2014;64:e1-76. [Crossref] [PubMed]
- Engelmann MD, Svendsen JH. Inflammation in the genesis and perpetuation of atrial fibrillation. Eur Heart J 2005;26:2083-92. [Crossref] [PubMed]
- Sharma G, Ghati N, Sharique M, et al. Role of inflammation in initiation and maintenance of atrial fibrillation in rheumatic mitral stenosis - An analytical cross-sectional study. J Arrhythm 2020;36:1007-15. [Crossref] [PubMed]
- Chung MK, Martin DO, Sprecher D, et al. C-reactive protein elevation in patients with atrial arrhythmias: inflammatory mechanisms and persistence of atrial fibrillation. Circulation 2001;104:2886-91. [Crossref] [PubMed]
- Wu N, Xu B, Xiang Y, et al. Association of inflammatory factors with occurrence and recurrence of atrial fibrillation: a meta-analysis. Int J Cardiol 2013;169:62-72. [Crossref] [PubMed]
- Marcus GM, Smith LM, Ordovas K, et al. Intracardiac and extracardiac markers of inflammation during atrial fibrillation. Heart Rhythm 2010;7:149-54. [Crossref] [PubMed]
- Li J, Solus J, Chen Q, et al. Role of inflammation and oxidative stress in atrial fibrillation. Heart Rhythm 2010;7:438-44. [Crossref] [PubMed]
- Attar A, Marzban P, Moaref A, et al. The association of plasma high-sensitivity C-reactive protein level with rheumatic heart disease: The possible role of inflammation. Indian Heart J 2018;70:346-9. [Crossref] [PubMed]
- Sharma G, Shetkar S, Bhasin A, et al. High sensitive C-reactive protein and interleukin 6 in atrial fibrillation with rheumatic mitral stenosis from Indian cohort. Indian Heart J 2017;69:505-11. [Crossref] [PubMed]
- Shiba M, Sugano Y, Ikeda Y, et al. Presence of increased inflammatory infiltrates accompanied by activated dendritic cells in the left atrium in rheumatic heart disease. PLoS One 2018;13:e0203756 [Crossref] [PubMed]
- Zhang YL, Teng F, Han X, et al. Selective blocking of CXCR2 prevents and reverses atrial fibrillation in spontaneously hypertensive rats. J Cell Mol Med 2020;24:11272-82. [Crossref] [PubMed]
- Hohmann C, Pfister R, Mollenhauer M, et al. Inflammatory cell infiltration in left atrial appendageal tissues of patients with atrial fibrillation and sinus rhythm. Sci Rep 2020;10:1685. [Crossref] [PubMed]
- Yamashita T, Sekiguchi A, Iwasaki YK, et al. Recruitment of immune cells across atrial endocardium in human atrial fibrillation. Circ J 2010;74:262-70. [Crossref] [PubMed]
- Young-Xu Y, Jabbour S, Goldberg R, et al. Usefulness of statin drugs in protecting against atrial fibrillation in patients with coronary artery disease. Am J Cardiol 2003;92:1379-83. [Crossref] [PubMed]
- Pfister R, Michels G, Brägelmann J, et al. Plasma vitamin C and risk of hospitalisation with diagnosis of atrial fibrillation in men and women in EPIC-Norfolk prospective study. Int J Cardiol 2014;177:830-5. [Crossref] [PubMed]
- Hemilä H, Suonsyrjä T. Vitamin C for preventing atrial fibrillation in high risk patients: a systematic review and meta-analysis. BMC Cardiovasc Disord 2017;17:49. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods 2015;12:453-7. [Crossref] [PubMed]
- Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882-3. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Doncheva NT, Morris JH, Gorodkin J, et al. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res 2019;18:623-32. [Crossref] [PubMed]
- Friendly M. Corrgrams: Exploratory Displays for Correlation Matrices. The American Statian 2002;56:316-24. [Crossref]
- Chugh SS, Havmoeller R, Narayanan K, et al. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation 2014;129:837-47. [Crossref] [PubMed]
- Magalhães L, Figueiredo M, Cintra FD, et al. II Diretrizes Brasileiras de Fibrilação Atrial. Arquivos Brasilros de Cardiologia 2016;106.
- Chugh SS, Blackshear JL, Shen WK, et al. Epidemiology and natural history of atrial fibrillation: clinical implications. J Am Coll Cardiol 2001;37:371-8. [Crossref] [PubMed]
- January CT, Wann LS, Calkins H, et al. 2019 AHA/ACC/HRS Focused Update of the 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society in Collaboration With the Society of Thoracic Surgeons. Circulation 2019;140:e125-51. [Crossref] [PubMed]
- Hu YF, Chen YJ, Lin YJ, et al. Inflammation and the pathogenesis of atrial fibrillation. Nat Rev Cardiol 2015;12:230-43. [Crossref] [PubMed]
- Hagiwara Nobuhisa. Inflammation and atrial fibrillation. Circulation Journal 2010;74:246-7. [Crossref] [PubMed]
- Katritsis DG. Is atrial fibrillation an inflammatory disorder? Eur Heart J 2006;27:886-author reply 886. [Crossref] [PubMed]
- Liu Y, Shi Q, Ma Y, et al. The role of immune cells in atrial fibrillation. J Mol Cell Cardiol 2018;123:198-208. [Crossref] [PubMed]
- Qu YC, Du YM, Wu SL, et al. Activated nuclear factor-kappaB and increased tumor necrosis factor-alpha in atrial tissue of atrial fibrillation. Scand Cardiovasc J 2009;43:292-7. [Crossref] [PubMed]
- Luan Y, Guo Y, Li S, et al. Interleukin-18 among atrial fibrillation patients in the absence of structural heart disease. Europace 2010;12:1713-8. [Crossref] [PubMed]
- Psychari SN, Apostolou TS, Sinos L, et al. Relation of elevated C-reactive protein and interleukin-6 levels to left atrial size and duration of episodes in patients with atrial fibrillation. Am J Cardiol 2005;95:764-7. [Crossref] [PubMed]
- Marcus GM, Whooley MA, Glidden DV, et al. Interleukin-6 and atrial fibrillation in patients with coronary artery disease: data from the Heart and Soul Study. Am Heart J 2008;155:303-9. [Crossref] [PubMed]
- Liuba I, Ahlmroth H, Jonasson L, et al. Source of inflammatory markers in patients with atrial fibrillation. Europace 2008;10:848-53. [Crossref] [PubMed]
- Wang CH, Hu DY, Tang CZ, et al. Changes of interleukin-1beta and tumor necrosis factor-alpha of right atrial appendages in patients with rheumatic valvular disease complicated with chronic atrial fibrillation. Zhonghua Xin Xue Guan Bing Za Zhi 2005;33:522-5. [PubMed]
- Zhao F, Zhang S, Shao Y, et al. Calreticulin overexpression correlates with integrin-α5 and transforming growth factor-β1 expression in the atria of patients with rheumatic valvular disease and atrial fibrillation. Int J Cardiol 2013;168:2177-85. [Crossref] [PubMed]
- Verheule S, Sato T, Everett T, et al. Increased vulnerability to atrial fibrillation in transgenic mice with selective atrial fibrosis caused by overexpression of TGF-beta1. Circ Res 2004;94:1458-65. [Crossref] [PubMed]
- Chang SH, Yeh YH, Lee JL, et al. Transforming growth factor-β-mediated CD44/STAT3 signaling contributes to the development of atrial fibrosis and fibrillation. Basic Res Cardiol 2017;112:58. [Crossref] [PubMed]
- Kunamalla A, Ng J, Parini V, et al. Constitutive Expression of a Dominant-Negative TGF-β Type II Receptor in the Posterior Left Atrium Leads to Beneficial Remodeling of Atrial Fibrillation Substrate. Circ Res 2016;119:69-82. [Crossref] [PubMed]
- Lindhardsen J, Ahlehoff O, Gislason GH, et al. Risk of atrial fibrillation and stroke in rheumatoid arthritis: Danish nationwide cohort study. BMJ 2012;344:e1257 [Crossref] [PubMed]
- Kreidberg JA, Sariola H, Loring JM, et al. WT-1 is required for early kidney development. Cell 1993;74:679-91. [Crossref] [PubMed]
- Wang L, Zhao XC, Cui W, et al. Genetic and Pharmacologic Inhibition of the Chemokine Receptor CXCR2 Prevents Experimental Hypertension and Vascular Dysfunction. Circulation 2016;134:1353-68. [Crossref] [PubMed]
- Wang L, Zhang YL, Lin QY, et al. CXCL1-CXCR2 axis mediates angiotensin II-induced cardiac hypertrophy and remodelling through regulation of monocyte infiltration. Eur Heart J 2018;39:1818-31. [Crossref] [PubMed]
- Paradis P, Schiffrin EL. CXCL1-CXCR2 lead monocytes to the heart of the matter. Eur Heart J 2018;39:1832-4. [Crossref] [PubMed]
- Zhang YL, Cao HJ, Han X, et al. Chemokine Receptor CXCR-2 Initiates Atrial Fibrillation by Triggering Monocyte Mobilization in Mice. Hypertension 2020;76:381-92. [Crossref] [PubMed]
- Stankiewicz AM, Goscik J, Swiergiel AH, et al. Social stress increases expression of hemoglobin genes in mouse prefrontal cortex. BMC Neurosci 2014;15:130. [Crossref] [PubMed]
- Hristov M, Zernecke A, Bidzhekov K, et al. Importance of CXC chemokine receptor 2 in the homing of human peripheral blood endothelial progenitor cells to sites of arterial injury. Circ Res 2007;100:590-7. [Crossref] [PubMed]
- Majumdar S, Gonder D, Koutsis B, et al. Characterization of the human beta-thromboglobulin gene. Comparison with the gene for platelet factor 4. J Biol Chem 1991;266:5785-9. [Crossref] [PubMed]
- Maneerat Y, Prasongsukarn K, Benjathummarak S, et al. PPBP and DEFA1/DEFA3 genes in hyperlipidaemia as feasible synergistic inflammatory biomarkers for coronary heart disease. Lipids Health Dis 2017;16:80. [Crossref] [PubMed]
- Song ZY, Wang F, Cui SX, et al. Knockdown of CXCR4 Inhibits CXCL12-Induced Angiogenesis in HUVECs through Downregulation of the MAPK/ERK and PI3K/AKT and the Wnt/β-Catenin Pathways. Cancer Invest 2018;36:10-8. [Crossref] [PubMed]
- Vila-Coro AJ, Rodríguez-Frade JM, Martín De Ana A, et al. The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J 1999;13:1699-710. [Crossref] [PubMed]
- Tian Y, Yin H, Deng X, et al. CXCL12 induces migration of oligodendrocyte precursor cells through the CXCR4-activated MEK/ERK and PI3K/AKT pathways. Mol Med Rep 2018;18:4374-80. [Crossref] [PubMed]
- Torossian F, Anginot A, Chabanon A, et al. CXCR7 participates in CXCL12-induced CD34+ cell cycling through β-arrestin-dependent Akt activation. Blood 2014;123:191-202. [Crossref] [PubMed]
- Hu XM, Zhang H, Xu H, et al. Chemokine receptor CXCR4 regulates CaMKII/CREB pathway in spinal neurons that underlies cancer-induced bone pain. Sci Rep 2017;7:4005. [Crossref] [PubMed]
- Sjaarda J, Gerstein H, Chong M, et al. Blood CSF1 and CXCL12 as Causal Mediators of Coronary Artery Disease. J Am Coll Cardiol 2018;72:300-10. [Crossref] [PubMed]
- Mehta NN, Li M, William D, et al. The novel atherosclerosis locus at 10q11 regulates plasma CXCL12 levels. Eur Heart J 2011;32:963-71. [Crossref] [PubMed]
- Tavakolian Ferdousie V, Mohammadi M, Hassanshahi G, et al. Serum CXCL10 and CXCL12 chemokine levels are associated with the severity of coronary artery disease and coronary artery occlusion. Int J Cardiol 2017;233:23-8. [Crossref] [PubMed]
- von Stebut E, Metz M, Milon G, et al. Early macrophage influx to sites of cutaneous granuloma formation is dependent on MIP-1alpha /beta released from neutrophils recruited by mast cell-derived TNFalpha. Blood 2003;101:210-5. [Crossref] [PubMed]
- Barczyk A, Pierzchała E, Caramori G, Sozańska E. Increased expression of CCL4/MIP-1β in CD8+ cells and CD4+ cells in sarcoidosis. Int J Immunopathol Pharmacol 2014;27:185-93. [Crossref] [PubMed]
- Mrugacz M. CCL4/MIP-1beta levels in tear fluid and serum of patients with cystic fibrosis. J Interferon Cytokine Res 2010;30:509-12. [Crossref] [PubMed]
- Vilá LM, Molina MJ, Mayor AM, et al. Association of serum MIP-1alpha, MIP-1beta, and RANTES with clinical manifestations, disease activity, and damage accrual in systemic lupus erythematosus. Clin Rheumatol 2007;26:718-22. [Crossref] [PubMed]
- Dai YJ, Li YY, Zeng HM, et al. Effect of pharmacological intervention on MIP-1α, MIP-1β and MCP-1 expression in patients with psoriasis vulgaris. Asian Pac J Trop Med 2014;7:582-4. [Crossref] [PubMed]
- Zacharias A, Schwann TA, Riordan CJ, et al. Obesity and risk of new-onset atrial fibrillation after cardiac surgery. Circulation 2005;112:3247-55. [Crossref] [PubMed]
- Korantzopoulos P, Kolettis TM. Obesity and the risk of new-onset atrial fibrillation. JAMA 2005;293:1974-author reply 1975. [PubMed]
- Mathews JA, Wurmbrand AP, Ribeiro L, et al. Induction of IL-17A Precedes Development of Airway Hyperresponsiveness during Diet-Induced Obesity and Correlates with Complement Factor D. Front Immunol 2014;5:440. [Crossref] [PubMed]
- Chen Y, Chang G, Chen X, et al. IL-6-miR-210 Suppresses Regulatory T Cell Function and Promotes Atrial Fibrosis by Targeting Foxp3. Mol Cells 2020;43:438-47. [PubMed]
(English Language Editor: R. Scott)


