
A phase II study of everolimus in patients with advanced solid malignancies with TSC1, TSC2, NF1, NF2 or STK11 mutations
Introduction
Mammalian target of rapamycin (mTOR) is a key protein kinase that regulates cell growth, proliferation and survival (1). Activation of the mTOR pathway has been implicated in the development of several malignancies (2,3). This protein kinase is mainly activated via the PI3K pathway through AKT and the tuberous sclerosis complex (TSC1/TSC2) (1). This pathway is also regulated by tumor suppressors such as STK11 and NF1, which are frequently altered across different cancers (4,5). STK11, also known as LKB1, activates mTOR through activation of AMPK and phosphorylation of TSC2, whereas NF1 prevents the downstream activation of the mTOR pathway by terminating the active state of RAS proteins (4,5). Mutations in TSC1, TSC2, STK11 and NF1 genes can lead to mTOR pathway dysregulation and promote tumor cell growth (6). Inhibiting mTOR could therefore represent an approach to treat solid tumors that harbor mutations in tumor suppressors such as STK11, NF1, TSC1 and TSC2.
Inhibitors of the mTOR pathway have demonstrated activity in different human solid tumor cell lines and in mouse xenograft models (7-10). Everolimus is an mTOR inhibitor that was first approved in 2009 by the the U.S. Food and Drug Administration (FDA) for the treatment of patients with advanced renal cell carcinoma (RCC) (11). Thereafter, approvals for everolimus for the treatment of different solid tumors were granted including subependymal giant cell astrocytoma, advanced hormone receptor positive HER-2 negative breast cancer, advanced pancreatic neuroendocrine and GI cancers and lung cancers, renal cell carcinoma, renal angiomyolipoma and tuberous sclerosis complex (12). Similarly other mTOR inhibitors such as temsirolimus, have also been studied in multiple cancers, and temsirolimus is currently approved in patients with renal cell carcinoma (13). Nevertheless, despite showing promising activity in preclinical studies across a variety of different cancers known to upregulate mTOR signaling, drugs such as everolimus and temsirolimus have failed to demonstrate significant efficacy in clinical trials—possibly owing to the lack of strong biomarkers that predict for responsiveness to mTOR inhibition (14).
In an attempt to identify predictors of response to everolimus, Iyer et al. performed whole genome sequencing (WGS) on tumoral DNA from a patient with metastatic bladder cancer that achieved complete response while on everolimus, and identified a two-base-pair deletion in the TSC1 gene and a nonsense mutation in the NF2 gene (15). Additional analyses by Iyer and colleagues also found a longer duration of response with everolimus in patients whose tumors harbored TSC1 mutations, compared to patients with wild type (WT) (15). Additionally, findings from other groups suggest a role for mTOR inhibition in malignant peripheral nerve sheath tumors (MPNSTs) with NF1 mutation, and breast cancer with STK11 mutation (16,17). Based on these observations, we designed a phase II basket study to examine the activity of mTOR inhibition in patients with solid tumors containing TSC1, TSC2, NF1, NF2, or STK11 mutations. Everolimus was selected as the mTOR inhibitor of choice for this study given its relatively well tolerated toxicity profile and ease of oral administration.
We present the following article in accordance with the TREND reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-195).
Methods
Study design
This was a single-arm, open label, single-institution phase II basket study of everolimus in patients with advanced solid tumors (ClinicalTrials.gov identifier NCT02352844) conducted at the outpatient oncology clinics at Washington University School of Medicine and Alvin J. Siteman Cancer Center in St. Louis, MO. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Washington University Human Research Protection Office institutional review board (IRB ID # 201502067). All patients provided written informed consent for participation in this study.
Eligibility
Adults (age ≥18 years) with histologically confirmed diagnosis of advanced solid tumor (metastatic, recurrent or unresectable) with mutations in TSC1, TSC2, NF1, NF2 or STK11 genes, who had failed at least one line of standard of care systemic therapy were eligible. Patients needed to have a 2 week washout between the last line of antineoplastic therapy and initiation of everolimus. There was no limit to the number of prior treatments patients could have received prior to enrollment. Further eligibility criteria included adequate bone marrow function (leukocytes >3,000/mcl, ANC >1,500/mcL, platelets >100,000 and hemoglobin >9.0 g/dL), hepatic function (total serum bilirubin ≥2.0× normal limit and AST/ALT ≤2.5× normal limit), and renal function (serum creatinine ≤1.5× normal limit or creatinine clearance 45mL/min), fasting cholesterol ≤300 mg/dL and fasting triglycerides ≤2.5× normal limit. Patients were required to have an ECOG performance status of 0–2. Patients with symptomatic brain metastases and those on chronic treatment with corticosteroids, or other immunosuppressive agents, were excluded.
Mutations in TSC1, TSC2, NF1, NF2 or STK11 genes were assessed using in-house and/or commercially available targeted-next generation sequencing (NGS) assays that utilized DNA extracted from formalin fixed paraffin embedded (FFPE) tissue blocks as part of their standard of care treatment (18). Patients with circulating tumor DNA (ctDNA) test results demonstrating mutations in these genes were also eligible for enrollment (19).
Treatment procedures
All eligible patients were treated with everolimus at an initial dose of 10 mg orally daily as a take-home medication in cycles of 28 days. Treatment with everolimus was continued until disease progression or unacceptable toxicity, whichever occurred first. Everolimus-related toxicities were managed with temporary dose delays or subsequent dose reductions. Dose reductions of 50% were implemented for recurrent grade 2 stomatitis, grade 3 hyperlipidemia or grade 3 hyperglycemia, and for grade 2 non-infectious pneumonitis. The study drug was discontinued if more than two dose reductions (50% and then 75%) were required due to toxicities. The use of live vaccinations was not permitted within a week of starting study treatment and during the study.
Efficacy and safety
Patients were evaluated clinically and by laboratory assessment with every cycle of everolimus in the outpatient clinic (every 4 weeks). Baseline scans were followed by repeat imaging with computed tomography (CT) scans obtained every 8 weeks (2 cycles) to evaluate response to treatment. Tumor response and progression were defined by the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 (20). The duration of overall response was defined as the time from when the measurement criteria for complete response (CR) or partial response (PR), were first met, until the time at which recurrent or progressive disease was objectively documented. Duration of stable disease (SD) was similarly defined. Patients who discontinued study therapy prior to the first radiological exam were not considered evaluable for response, but were still considered evaluable for toxicity. Adverse events (AEs) were recorded and graded according to the National Cancer Institute Common Terminology Criteria version 4.0 (21).
Outcomes
The primary end point of this study was overall response rate (ORR). Secondary end points included an examination of the correlation between presence of different mutations in the mTOR pathway and response to everolimus, and investigation of the genetic changes associated with disease progression following initial response to everolimus.
Statistical analysis
The study was designed to assess the response rate of everolimus in solid tumors that harbor TSC1, TSC2, NF1, NF2 and STK11 mutations. Since this was a study in patients with specifically targeted mutations, a substantial treatment effect size was expected. Based on historical data, we assumed a null hypothesis of an ORR of <1% in this setting. We hypothesized that an overall response rate of 30% or higher would warrant further investigation. Assuming a dropout rate near to zero, a target accrual of 12 patients was planned for this study, based on the exact single-stage design with 85% power at a 1-sided 0.05 significance level (using ph2single function from library clinfun in the R package) (22,23).
All data including demographic and clinical characteristics of enrolled patients, and response and toxicity by grade were summarized using descriptive statistics, i.e., statistics for continuous variables included medians and ranges, and statistics for qualitative variables included counts and percentages. Waterfall plot was graphed to visualize tumor responses in terms of percent change of tumor size from baseline.
Results
Patient characteristics
Between February 2016 and June 2017, 12 patients with advanced solid malignancies were enrolled to this study. Patient characteristics are listed in Table 1. The majority of patients had a diagnosis of non-small cell lung cancer (NSCLC) (6/12; 50%), followed by carcinoma of unknown primary site (2/12;17%), small cell lung cancer (1/12; 8.3%), gall bladder adenocarcinoma (1/12; 8.3%), pancreatic adenocarcinoma (1/12; 8.3%) and cervical cancer (1/12; 8.3%). Seven patients (58%) were female, and eleven (92%) were Caucasian. The median age of patients enrolled to the trial was 66 years (range, 40–86 years). With regard to evaluable patients (8/12; 75%), four patients had STK11 mutations and four harbored mutations in NF1.

Full table
Antitumor activity
Four patients were not evaluable for response due to study withdrawal prior to first evaluation. Three of these four patients were lost to follow-up and one patient withdrew from study after 2 weeks at the discretion of the treating provider. The median number of cycles administered was 2 across the cohort of evaluable patients. Among the eight evaluable patients, one complete response (CR) was observed in a patient with non-small cell lung cancer representing a response rate of 12.5%. This patient only received treatment with everolimus for 2 weeks, before discontinuation due to treatment-related toxicities including grade 3 hyponatremia and a grade 4 pericardial effusion. However, follow-up assessment at 10 week mark showed a complete response that was durable and lasted more than 10 months. One patient with metastatic NSCLC (12.5%) demonstrated SD and 6 (75%) had disease progression (Figure 1) on the first post-treatment scan. The patient with SD was on therapy for 6 cycles before discontinuing treatment for progression. Notably, in this small cohort, disease control (CR, PR and SD) was observed in 2 of the 5 patients with lung adenocarcinoma.
Safety
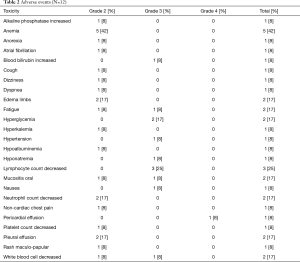
Table 2 summarizes grade 2 and higher treatment-related toxicities. Twelve patients that received everolimus were evaluable for toxicity. Overall, treatment with everolimus was well tolerated and no new toxicities were observed. Grade 2 toxicities observed in greater than 10% patients included anemia in 5 patients (42%), peripheral edema in 2 patients (17%), neutrophil decrease in 2 patients (17%), and pleural effusion in 2 patients (17%). Grade 3 toxicities included lymphopenia in 3 patients (25%), hyperglycemia in 2 patients (17%), followed by fatigue, increased blood bilirubin, hypertension, hyponatremia, oral mucositis, nausea, and low white blood cell count in 1 patient each (8% each). Grade 4 pericardial effusions were observed in 2 patients on study. However, only one of these events was assessed as being related to everolimus and led to treatment discontinuation.
Full table
Molecular characteristics
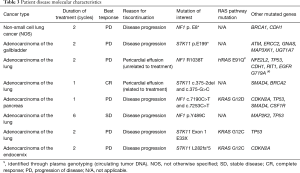
Of the patients who were evaluable for a response, 50% (n=4) of the patients had tumors with NF1 mutations and 50% (n=4) had tumors with STK11 mutations (Table 3). None of the alterations in NF1 or STK11 were recurrent. Mutations observed in NF1 were missense (n=3) and nonsense (n=1), and STK11 mutations were mostly nonsense (n=2). The single patient with a CR had two noncoding variants in cis in the STK11 gene, with one of the alterations occurring at a canonical splice acceptor site of the second intron, and the second occurring upstream of this splice site acceptor site. Tumor from the patient with SD demonstrated a missense mutation in NF1.
Full table
Discussion
This phase II study explored the response rate to everolimus in a heavily pre-treated population of patients with solid tumors with mutations in tumor suppressor genes that regulate the mTOR pathway. Overall, this study failed to demonstrate significant single agent activity with everolimus in solid tumors with mutations in NF1, NF2, TSC1, TSC2, and STK11. Nevertheless, our findings suggest that everolimus may be active in a subset of patients including those with adenocarcinoma of the lung. Two of four evaluable patients with lung adenocarcinoma demonstrated disease control with everolimus in our cohort. It is possible that the lack of activity with everolimus in other tumor types included in our study could be a result of differences in tumor biology associated with site of origin. Such differential responses to targeted therapies based on tissue of origin have previously been reported in the context of BRAF and KRAS inhibitors (24-26). For instance, while inhibitors targeting the BRAF V600E mutation or KRAS G12C mutation have demonstrated considerable activity in melanoma (24) and lung cancers (25), respectively, these drugs have failed to demonstrate significant single agent activity in patients with colorectal cancer (26).
Alternatively, the lack of efficacy with everolimus in this study, could also be related to the effect of co-occurring genomic alterations in tumor samples. Patients that derived benefit from everolimus in our study had lung adenocarcinomas that contained NF1 and STK11 mutations, while lacking mutations in the RAS pathway (such as KRAS and HRAS). On the contrary, the lung adenocarcinoma patients whose disease failed to respond to treatment, demonstrated combined alteration of KRAS and STK11 or NF1 and HRAS. Furthermore, two other patients with non-lung malignancies who failed to respond to treatment, also had tumors with co-existing KRAS mutations, raising the possibility that co-occurring KRAS mutations could predict for resistance to everolimus in this patient population (Table 3). Such a hypothesis would be supported by observations from pre-clinical studies in which the presence of KRAS mutations has been shown to predict for resistance to mTOR inhibition in PIK3CA mutated cancer cell lines and xenografts (27). In this study, Di Nicolantonio et al. observed that colorectal cancer cell lines carrying PIK3CA mutations were responsive to everolimus, except when KRAS mutations occurred concomitantly (27). Furthermore, when they performed genetic analysis of tumors from patients with different solid tumors (mostly colorectal and breast cancer) that were treated with everolimus, patients whose tumors carried PIK3CA and KRAS mutations displayed lack of clinical benefit from everolimus (27). Finally, the functional significance of the specific mutations affecting genes of interest in our study was not always clear—particularly in samples that contained missense mutations. It is possible that some of the variants observed in this study were passenger mutations that did not lead to activation of mTOR signaling.
Overall, our findings suggest that patients with adenocarcinoma of the lung with NF1 or STK11 mutations without co-occurring mutations in KRAS or HRAS genes may derive some benefit from everolimus (Figure 2). STK11 and NF1 mutations are frequently seen in lung cancer (17.4% and 11.7%, respectively) (28,29). Nevertheless, these observations should be viewed as hypothesis generating and will require further validation in larger studies. Additional pre-clinical studies exploring genomic features that predict for response to everolimus in lung adenocarcinoma will also be necessary to inform the design of future studies.
Acknowledgments
Funding: Funding for this investigator-initiated study was provided by Novartis Pharmaceuticals.
Footnote
Reporting Checklist: The authors have completed the TREND reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-195
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-195
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-195). Dr. BP receives research support from Bristol Myers Squibb. Dr. BP has received speaker honoraria from BioAscend and OncLive, has serve as a consultant for AstraZeneca and Guidepoint, and has received assay supply by Roche.Roche for laboratory research outside the submitted work. Mr. LV reports an unpaid consulting agreement with Jounce Therapeutics, Inc. Dr. HP reports research funding to her institution from Ambrx, Amgen, Aprea Therapeutics AB, Array BioPharma, Bayer, BeiGene, BJ Bioscience, Bristol-Myers Squibb, Daiichi Sankyo, Eli Lilly, Elicio Therapeutics, EMD Serono, Genentech, Gilead Sciences, GlaxoSmithKline, Gossamer Bio, Hoffman-LaRoche, Hutchison Medi-Pharma, ImmuneOncia Therapeutics, Incyte, Jounce Therapeutics, Macrogenics, MedImmune, Medivation, Merck, Millenium, Mirati Therapeutics, Novartis, Oncologie, Pfizer, PsiOxus Pharmaceuticals, Seattle Genetics, Synermore Biologics, Puma Biotechnology, Regeneron, Taiho, Vertex, Turning Point Therapeutics, Xencor, Inc, Genentech, BJ Bioscience, Vedanta Bioscences, TopAlliance Biosciences outside this submitted work. Dr. DM has served on Advisory Boards for Boehringer-Ingelheim, Abbvie, Celgene, Takeda and PharmaMar outside of this submitted work. Dr. DM serves as an unpaid editorial board member of Journal of Thoracic Disease from Sep 2020 to Aug 2022. Dr. RG has served on advisory boards for AbbVie, Inivata, Pfizer, Genentech, Bristol-Myers Squibb, Roche, Nektar, Merck, Celgene, Partner Therapeutics, GlaxoSmithKline, Jounce, and Achilles and has served as a consultant for AbbVie, Eli Lilly and Company, AstraZeneca, Pfizer, Genentech, Millennium Pharm, F. Hoffmann-La Roche, NeoHealth, Janssen, Amgen, and GenePlus. Dr. PP reports personal fees from Bristol Myers, personal fees from Merck, personal fees from Eisai, outside the submitted work. Dr. MQB has served as site PI for studies funded by Medimmune, AstraZeneca, Millenium, CRAB, Bristol Myers Squibb, and Alliance for Clinical Trials in Oncology, outside the submitted work. She also is on the Board of Directors for Lung Cancer Connection and serves on the American Society for Clinical Oncology e-Learning Editorial Board. Dr. SNW reports that the trial is supported by Novartis Pharmaceuticals. She reports funding from the SWOG-Clinical Trials Partnership, honoraria from the American Society for Clinical Oncology (ASCO) and serves as Chair of Data Safety Monitoring Board for a study through the Hoosier Oncology Group outside this submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. he study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Washington University Human Research Protection Office institutional review board (IRB ID # 201502067). All patients provided written informed consent for participation in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 2006;6:729-34. [Crossref] [PubMed]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell 2000;103:253-62. [Crossref] [PubMed]
- Wislez M, Spencer ML, Izzo JG, et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res 2005;65:3226-35. [Crossref] [PubMed]
- Gill RK, Yang SH, Meerzaman D, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene 2011;30:3784-91. [Crossref] [PubMed]
- Redig AJ, Capelletti M, Dahlberg SE, et al. Clinical and Molecular Characteristics of NF1-Mutant Lung Cancer. Clin Cancer Res 2016;22:3148-56. [Crossref] [PubMed]
- Vandekerkhove G, Todenhofer T, Annala M, et al. Circulating Tumor DNA Reveals Clinically Actionable Somatic Genome of Metastatic Bladder Cancer. Clin Cancer Res 2017;23:6487-97. [Crossref] [PubMed]
- Ma BB, Lui VW, Hui EP, et al. The activity of mTOR inhibitor RAD001 (everolimus) in nasopharyngeal carcinoma and cisplatin-resistant cell lines. Invest New Drugs 2010;28:413-20. [Crossref] [PubMed]
- Huynh H, Chow KH, Soo KC, et al. RAD001 (everolimus) inhibits tumour growth in xenograft models of human hepatocellular carcinoma. J Cell Mol Med 2009;13:1371-80. [Crossref] [PubMed]
- Chu C, Noel-Hudson MS, Boige V, et al. Therapeutic efficiency of everolimus and lapatinib in xenograft model of human colorectal carcinoma with KRAS mutation. Fundam Clin Pharmacol 2013;27:434-42. [Crossref] [PubMed]
- Liakakos T, Roukos DH. Everolimus and sunitinib: from mouse models to treatment of pancreatic neuroendocrine tumors. Future Oncol 2011;7:1025-9. [Crossref] [PubMed]
- Motzer RJ, Escudier B, Oudard S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet 2008;372:449-56. [Crossref] [PubMed]
- Affinitor (everolimus) package insert.
- Hudes G, Carducci M, Tomczak P, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med 2007;356:2271-81. [Crossref] [PubMed]
- Tian T, Li X, Zhang J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int J Mol Sci 2019;20:755. [Crossref] [PubMed]
- Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 2012;338:221. [Crossref] [PubMed]
- Johannessen CM, Reczek EE, James MF, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A 2005;102:8573-8. [Crossref] [PubMed]
- Parachoniak CA, Rankin A, Gaffney B, et al. Exceptional durable response to everolimus in a patient with biphenotypic breast cancer harboring an STK11 variant. Cold Spring Harb Mol Case Stud 2017;3:a000778 [Crossref] [PubMed]
- Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015;121:631-9. [Crossref] [PubMed]
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res 2019;25:4691-700. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Common Terminology Criteria for Adverse Events (CTCAE) version 4.0.
- A'Hern RP. Sample size tables for exact single-stage phase II designs. Stat Med 2001;20:859-66. [Crossref] [PubMed]
- Khan I, Sarker SJ, Hackshaw A. Smaller sample sizes for phase II trials based on exact tests with actual error rates by trading-off their nominal levels of significance and power. Br J Cancer 2012;107:1801-9. [Crossref] [PubMed]
- Dummer R, Ascierto PA, Gogas HJ, et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2018;19:1315-27. [Crossref] [PubMed]
- Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020;383:1207-17. [Crossref] [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [Crossref] [PubMed]
- Di Nicolantonio F, Arena S, Tabernero J, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest 2010;120:2858-66. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- . Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]




