
Genomic alteration profiles of lung cancer and their relationship to clinical features and prognosis value using individualized genetic testing
Introduction
Genetic testing using the next-generation sequencing (NGS) analysis is becoming increasingly recommended for cancer diagnosis and treatment decision-making (1,2). Traditionally, the NGS technique was applied to tumor tissues to examine gene alterations related to specific cancers (3,4). Currently, an increasing number of medical centers have recommended the liquid biopsy technique to admitted patients, owing to its advantages of minimal invasion and ability to probe the genomic profile of tumors in real time (5-7). In China, the choice between different commercial genetic testing products to conduct sequencing based on blood samples is available; for many tumor departments, clinical data accompanied by genetic testing results have been accumulated in recent years. A timely analysis of the data features in the real world would greatly push forward the progress of understanding the onset and development of cancer.
NGS has been used to clarify the gene mutations differences of ctDNA in peripheral blood between early-stage lung cancer and benign nodules. RNF213 mutations is identified to be the biomarker of early-stage lung cancer (8). Targeted deep sequencing is also performed to compare the proportion of positive DNA mutations (40 mutations identified) in the four samples (primary tumors, pulmonary venous blood, peripheral blood, and rib bone marrow fluid) in 10 lung patients (9). However, the samples size is small in previous studies. The pulmonary venous blood is the most convenient way to detect the gene mutation. As the NGS technological development, the sensitivity to detect mutations in peripheral venous blood is increasing. Large sample research is needed to verify the efficiency of NGS on gene mutation in peripheral venous blood.
Among all malignancies, lung cancer is the leading cause of cancer-related death in China and worldwide. Focusing on lung cancer, some recognized genes (like TP53, KRAS and EGFR, etc.) have been widely surveyed for their roles in development and relapse. For example, many lung cancer patients carry mutated EGFR and are generally recommended targeted therapy. Despite the widely studied roles of genes such as EGFR, it remains challenging but rewarding to probe a panel of other gene alterations . To date, sufficient data have been accumulated, including the sequencing results and clinical characteristics of lung cancer patients. However, limited studies have analyzed their relationships and the prognostic role of the genetic testing profiles in depth. Therefore, we conducted this study and aimed to use a panel targeting 197 genes and 38 fusions to observe the features of gene variations in lung cancer patients, as well as their prognostic values. Compared with previous study the sample size was large and the panel content was larger. This study will provide more information on gene mutation in cancer genetic analysis. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-1031).
Methods
Patients
Patients admitted in our hospital between 2016 and 2017 were eligible for inclusion in this study. The inclusion criteria were as follows: (I) primary lung cancer patients; and (II) those who had received OseqTM-Drug (Beijing Genomics institution, Beijing) genetic testing. Patients were excluded based on the following criteria: (I) those unwilling to sign the informed consent; and (II) incompatibility in the follow-up data collection. Finally, 33 patients were enrolled. This cohort comprised 25 males and eight females, with a mean age 58.9 years old (range, 43–82). Subjects received the mutation examination of a panel including 197 genes and 38 gene fusions, as shown in Tables 1 and 2. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the ethics committee of the Fourth Hospital of Hebei Medical University (No.: 2021KY255). The informed consent was taken from all the patients.

Full table

Full table
Sample collection and genomic DNA extraction
The samples were collected and detected as soon as possible. Briefly, for each patient, a total of 5 mL peripheral venous blood was collected in Ethylene Diamine Tetraacetic Acid (EDTA) anticoagulation tubes and stored at 4 °C prior to treatment. The circulating tumor DNAs (ctDNAs) and genomic DNAs were extracted. Briefly, 2 mL of blood was centrifuged at 2,000 g for 10 min, red blood cells and buffy coat were discarded, and the plasma was centrifuged at 16,000 g for 10 min at 4 °C. A volume of 500 μL purified plasma was treated with the TIANamp Blood DNA Kit (Tiangen Biotech, China).
Genotyping
The pre-hybridization DNA sample was treated through terminal repair, linker ligation, and pre-polymerase chain reaction (PCR) amplification. Next, the DNA fragment containing the target region was specifically captured by the biotin-labeled DNA probe, followed by the post-PCR process. After hybridization, the sequencing reaction was carried out using the OseqTM-drug Individualized Gene Testing Kit (Beijing Genomics institution, Beijing). The sequencing information of each target region was acquired and exported. After compared to the normal human genome sequence, any variation will be documented. For each subject, an individualized report was generated, which included the mutant gene name, nucleotide change, heterozygosity, inheritance pattern, and mutation types.
Statistical analysis
Data were mainly expressed as a frequency, and Chi-square analysis of the frequency distribution was used to determine the correlation between two nominal variables. P<0.05 was considered statistically significant.
Results
Clinical features
In total, 43 subjects were initially enrolled, including 26 males (mean age 59.1 years old, 43–75) and 17 females (mean age 58.3 years old, 34–82). Three patients who had not clearly received a subtype determination through section pathological examination were excluded. Among the remaining 40 participants, there were 35 cases of adenocarcinoma (ADC) and five cases of proven squamous cell carcinoma (SCC). No other sub-types were observed in this study. At the time of diagnosis, there were four cases in stage I, one case at stage II, 10 cases at stage III, and 28 cases at stage IV. Among all 43 subjects, 33 showed lymph node metastases at the time of diagnosis.
Mutation features
Within all the targets in the panel, 32 genes were observed with mutations, which were ranked according to their mutation frequency (Table 3). As expected, the EGFR gene exhibited the highest mutation rate, with 46.5% subjects (20 cases) carrying mutations of this gene. Also, the well-known oncogene, TP53, ranked second in mutation frequency. Other common mutations included NF1, FBXW7, HGF, etc. We further counted the frequencies of individuals with different mutation gene numbers. Patients carrying only one mutant gene exhibited the highest proportion, with 13 patients (28.9%) carrying mutants in one gene. There were four patients (9.3%) with mutants in two genes, three patients (7.0%) with mutants in three genes, one (2.3%) with mutants in four genes, and interestingly, three patients (7.0%) with mutants in five genes. For some of the zero-mutation subjects, we further checked this conclusion using the biopsy samples from the tumor tissue and ensured no detectable mutations. This result implied that there are still a large proportion of lung cancer patients without recognized mutations in our panel.

Full table
Correlations between mutations and cancer states
Next, we analyzed the association between gene mutation and disease states, such as the stage, subtype, and metastasis at the time of diagnosis. There were no statistical relationships between mutations and tumor stage, sex, age, etc. However, pathological subtypes were associated with several gene mutations (Table 4): FLI1, IGF1R, and NOTCH1. A significant correlation between mutant genes and stage at diagnosis was observed (Table 5), however this requires further confirmation, as there was only one case in these mutations: AKT2, AR, STK11, VEGFA, HDAC6, and ASPSCR. Moreover, for the 33 patients with lymph node metastases at the time of diagnosis, no correlation between metastasis and gene polymorphism was found.
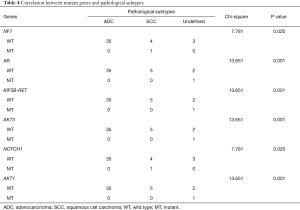
Full table
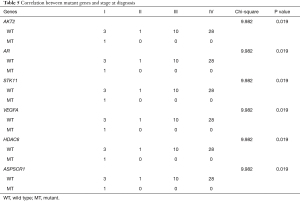
Full table
Correlations between gene mutations and prognosis
Furthermore, the prognostic value of mutation states was probed after the follow-up data collection. Relapse and later metastasis were surveyed regarding their relationship with gene mutations. There was no association between 1-year progression and gene mutations (Table 6). Similarly, there were no differences in the 1-year survival among patients with different gene mutations. Next, we compared the PFS and OS using the Kaplan-Meier (KM) method. To acquire a reliable conclusion, those gene mutations with only one positive case were not included. Namely, the following genes were analyzed: EGFR, TP53, MSH6, HER2, ZNF217, and BRCA1. None of the above gene mutations were found to be correlated with PFS. Moreover, OS was assessed, and none of the above six genes exhibited a relationship with OS. Taken together, gene mutation detection has a limited efficacy in the prediction of prognosis.

Full table
Discussion
In the present study, we applied a non-invasive method to detect peripheral blood DNA, and performed individualized genetic testing to survey the genomic alterations in lung cancer patients. NGS is a potential method in cancer research. NGS has many NGS has been shown to have a powerful effect on many aspects of lung cancer. NGS could detect the gene mutation (10), guide the application of sensitive drug (11), promote diagnosis (12) and prognosis (13) in lung cancer research. The mutant genes could be detected and they are associated with the tumorigenesis of lung cancer. In early stage of tumorigenesis, simultaneous mutation of EGFR and ALK could be induced through different tumor evolution (14). Further, many genes are mutant and differently expressed in lung cancer, the histone 3 lysine-27 demethylase KDM6A is associated with tumorigenesis and prognosis (15). COX1, COX2, COX3, ND1, ND2, ND4L, and ATP6 are abnormal increase in the plasma of non-small cell lung cancer (16). Thus, using NGS to clarify the mutant genes in peripheral blood is of great importance in lung cancer research.
In our high-throughput panel, we observed 32 genes with mutations. Also, approximately 18% of patients did not exhibit any mutation in either blood or tumor tissue samples, which may provide a different understanding of lung cancer onset. Moreover, we observed clear associations between gene mutations and cancer states at diagnosis.
Consistent with the widely established conclusion, EGFR mutation plays a prominent driving role in lung cancer onset (17,18). In our study, 46.5% of participants were diagnosed with EGFR mutation. These patients can largely benefit from the fluid biopsy followed by genetic testing, as targeted therapy can be selected for them using EGFR tyrosine kinase inhibitors (TKIs). For East Asians, EGFR exhibits the highest mutation frequency, and this ratio is lower in Americans/Europeans (19,20). TP53 ranked second in mutation frequency behind EGFR. This finding was seldomly noticed, as BRAF or KRAS, but not TP53, were generally among the most frequently observed mutations in lung cancer following EGFR. Our result further emphasized the tumor-suppressor role of TP53 in lung cancer development.
Interestingly, approximately 18% patients did not carry any known mutation. This may be due to the limited throughput of our testing panel, limited sensibility of the testing protocol, tumor heterogeneity, or some undiscovered mechanisms of lung cancer onset. We will pursue further research in the future, especially focusing on those cases without detectable mutations.
Based on our correlation analysis, some important genes were found to potentially influence the pathological subtypes. For example, SCC patients had higher mutation frequencies in FLI1, IGF1R, and NOTCH1, and a higher risk of lymph node metastasis. It is agreed that SCC exhibits increased metastasis and drug resistance, with a higher expression of markers being associated with lymph node metastasis (21-23). However, this is the first report linking FLI1, IGF1R, and NOTCH1 mutations with the lung SCC subtype. A previous report identified NOTCH1 or NOTCH2 mutations in 75% of cutaneous SCCs but in a lesser fraction of lung SCCs (24). Our findings provide a new insight into SCC development or transition.
However, the same gene may play different or even contrary roles in different lung cancer subtypes. For instance, in lung adenocarcinoma, Numb protein impairs tumor growth and inhibits the Notch pathway and epithelial-mesenchymal transition, whereas in lung squamous cell carcinoma, it may promote proliferation (25). In addition, we noticed that some mutant genes significantly influenced the metastasis/recurrence outcomes, especially ATM. Combined with metastasis at the time of diagnosis and in the post-treatment follow-up period, ATM mutation is the most frequent factor in all kinds of metastases. Published data supports that ATM sequence variants and expression could be used as a predictor for radiotherapy responses and chemotherapy resistance in breast cancer patients (26,27). Moreover, ATM mutation is an important driving factor of tumorigenesis (28-30). Following ATM, STK11 mutation was another important factor that repeatedly appeared in metastases. It was mentioned that inactivation of STK11 was closely related to tumor occurrence, dominantly in non-small cell lung cancer (31,32). Some scholars also claimed that STK11 mutation may also play a role in identifying thyroid carcinoma (33). Therefore, it is valuable to probe the detailed role of ATM and STK11 in suppressing tumor development and metastasis.
There are some limitations in the present work that should be noted. Firstly, the sample size was small, and many significances were derived from the crucial one or two individuals with multiple mutations. Also, due to the small sample size, our conclusions cannot be majorly applied in ADC and SCCs, and there is an obvious void with regards to small cell lung cancer. Furthermore, the follow-up period was so short that all the participants were still alive at the time of data analysis. Therefore, all of the results regarding the association between gene mutations and prognosis were negative. Moreover, the targets of the panel needed to be extended and the sensitivity requires enhancement, especially considering that 18% patients were negative in the mutation detection. So far, it is too early to ascertain whether the reason for this phenomenon lies in the testing means or the patients themselves. Lastly, this correlation study could not reveal the detailed mechanisms of some findings, especially those inconsistent with known reports (e.g., EGFR mutation was negatively correlated with lymph node metastases).
In summary, we evaluated the usefulness of tumor DNA sequencing from blood samples of lung cancer patients, which is a feasible and minimally invasive approach in cancer genetic analysis. EGFR and TP53 are the first two mutant genes; NF1, AR, KIF5B-RET, AKT1, AKT3 and NOTCH1 were associated with the pathological subtypes. AKT2, AR, STK11, VEGFA, HDAC6 and ASPSCR1 were associated with the stage at diagnosis. Although the usefulness of gene mutations in prognosis prediction is limited in this study, the mutant genes detected by NGS could also provide essential guide information for the diagnosis, prognosis and treatment drug selection lung cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-1031
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-1031
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-1031). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the ethics committee of the Fourth Hospital of Hebei Medical University (No.: 2021KY255).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sapari NS, Elahi E, Wu M, et al. Feasibility of low-throughput next generation sequencing for germline DNA screening. Clin Chem 2014;60:1549-57. [Crossref] [PubMed]
- Liu N, Kan J, Yu N, et al. Application of metagenomic next-generation sequencing technology for difficult lung lesions in patients with haematological diseases. Transl Cancer Res 2020;9:5245-54. [Crossref]
- Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016;7:65364-73. [Crossref] [PubMed]
- Barata PC, Koshkin VS, Funchain P, et al. Next-generation sequencing (NGS) of cell-free circulating tumor DNA and tumor tissue in patients with advanced urothelial cancer: a pilot assessment of concordance. Ann Oncol 2017;28:2458-63. [Crossref] [PubMed]
- Nakashima C, Sato A, Abe T, et al. Automated DNA extraction using cellulose magnetic beads can improve EGFR point mutation detection with liquid biopsy by efficiently recovering short and long DNA fragments. Oncotarget 2018;9:25181-92. [Crossref] [PubMed]
- Gornstein EL, Sandefur S, Chung JH, et al. BRCA2 Reversion Mutation Associated With Acquired Resistance to Olaparib in Estrogen Receptor-positive Breast Cancer Detected by Genomic Profiling of Tissue and Liquid Biopsy. Clin Breast Cancer 2018;18:184-8. [Crossref] [PubMed]
- Li T, Zheng Y, Sun H, et al. K-Ras mutation detection in liquid biopsy and tumor tissue as prognostic biomarker in patients with pancreatic cancer: a systematic review with meta-analysis. Med Oncol 2016;33:61. [Crossref] [PubMed]
- Jiang N, Zhou J, Zhang W, et al. RNF213 gene mutation in circulating tumor DNA detected by targeted next-generation sequencing in the assisted discrimination of early-stage lung cancer from pulmonary nodules. Thorac Cancer 2021;12:181-93. [Crossref] [PubMed]
- Goto T, Hirotsu Y, Oyama T, et al. Analysis of tumor-derived DNA in plasma and bone marrow fluid in lung cancer patients. Med Oncol 2016;33:29. [Crossref] [PubMed]
- Zhang S, Xu Y, Zhao P, et al. Integrated Analysis of Genomic and Immunological Features in Lung Adenocarcinoma With Micropapillary Component. Front Oncol 2021;11:652193 [Crossref] [PubMed]
- Nong L, Zhang Z, Xiong Y, et al. Comparison of next-generation sequencing and immunohistochemistry analysis for targeted therapy-related genomic status in lung cancer patients. J Thorac Dis 2019;11:4992-5003. [Crossref] [PubMed]
- Wan L, Liu Q, Liang D, et al. Circulating Tumor Cell and Metabolites as Novel Biomarkers for Early-Stage Lung Cancer Diagnosis. Front Oncol 2021;11:630672 [Crossref] [PubMed]
- Jin R, Peng L, Shou J, et al. EGFR-Mutated Squamous Cell Lung Cancer and Its Association With Outcomes. Front Oncol 2021;11:680804 [Crossref] [PubMed]
- Lee KC, Koh J, Chung DH, et al. A case of concomitant EGFR/ALK alteration against a mutated EGFR background in early-stage lung adenocarcinoma. J Pathol Transl Med 2021;55:139-44. [Crossref] [PubMed]
- Leng X, Wang J, An N, et al. Histone 3 lysine-27 demethylase KDM6A coordinates with KMT2B to play an oncogenic role in NSCLC by regulating H3K4me3. Oncogene 2020;39:6468-79. [Crossref] [PubMed]
- Wang L, Wang J, Jia E, et al. Plasma RNA sequencing of extracellular RNAs reveals potential biomarkers for non-small cell lung cancer. Clin Biochem 2020;83:65-73. [Crossref] [PubMed]
- Shin S, Kim J, Kim Y, et al. Assessment of real-time PCR method for detection of EGFR mutation using both supernatant and cell pellet of malignant pleural effusion samples from non-small-cell lung cancer patients. Clin Chem Lab Med 2017;55:1962-9. [Crossref] [PubMed]
- Takahama T, Sakai K, Takeda M, et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study). Oncotarget 2016;7:58492-9. [Crossref] [PubMed]
- Li S, Li L, Zhu Y, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer 2014;110:2812-20. [Crossref] [PubMed]
- Buder A, Hochmair MJ, Setinek U, Pirker R, Filipits M. EGFR mutation tracking predicts survival in advanced EGFR-mutated non-small cell lung cancer patients treated with osimertinib. Transl Lung Cancer Res 2020;9:239-45. [Crossref] [PubMed]
- Ren ZH, Zhang CP, Ji T. Expression of SOX2 in oral squamous cell carcinoma and the association with lymph node metastasis. Oncol Lett 2016;11:1973-9. [Crossref] [PubMed]
- Jiang R, Wang X, Jin Z, et al. Association of Nuclear PIM1 Expression with Lymph Node Metastasis and Poor Prognosis in Patients with Lung Adenocarcinoma and Squamous Cell Carcinoma. J Cancer 2016;7:324-34. [Crossref] [PubMed]
- Boelens MC, Kok K, van der Vlies P, et al. Genomic aberrations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer 2009;66:372-8. [Crossref] [PubMed]
- Wang NJ, Sanborn Z, Arnett KL, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A 2011;108:17761-6. [Crossref] [PubMed]
- Kikuchi H, Sakakibara-Konishi J, Furuta M, et al. Numb has distinct function in lung adenocarcinoma and squamous cell carcinoma. Oncotarget 2018;9:29379-91. [Crossref] [PubMed]
- Ho AY, Fan G, Atencio DP, et al. Possession of ATM sequence variants as predictor for late normal tissue responses in breast cancer patients treated with radiotherapy. Int J Radiat Oncol Biol Phys 2007;69:677-84. [Crossref] [PubMed]
- Knappskog S, Chrisanthar R, Løkkevik E, et al. Low expression levels of ATM may substitute for CHEK2 /TP53 mutations predicting resistance towards anthracycline and mitomycin chemotherapy in breast cancer. Breast Cancer Res 2012;14:R47. [Crossref] [PubMed]
- Jerzak KJ, Mancuso T, Eisen A. Ataxia-telangiectasia gene (ATM) mutation heterozygosity in breast cancer: a narrative review. Curr Oncol 2018;25:e176-80. [Crossref] [PubMed]
- Laitman Y, Boker-Keinan L, Berkenstadt M, et al. The risk for developing cancer in Israeli ATM, BLM, and FANCC heterozygous mutation carriers. Cancer Genet 2016;209:70-4. [Crossref] [PubMed]
- Thompson D, Duedal S, Kirner J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst 2005;97:813-22. [Crossref] [PubMed]
- Sanchez-Cespedes M, Parrella P, Esteller M, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res 2002;62:3659-62. [PubMed]
- Facchinetti F, Bluthgen MV, Tergemina-Clain G, et al. LKB1/STK11 mutations in non-small cell lung cancer patients: Descriptive analysis and prognostic value. Lung Cancer 2017;112:62-8. [Crossref] [PubMed]
- Wei S. STK11 Mutation Identified in Thyroid Carcinoma. Endocr Pathol 2016;27:65-9. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


