
Mesenchymal stem cells transfected with anti-miRNA-204-3p inhibit acute rejection after heart transplantation by targeting C-X-C motif chemokine receptor 4 (CXCR4) in vitro
Introduction
Heart transplantation (HTx) has been a promising treatment option for end-stage heart disorders for several decades (1). Undeniably, immune rejection is one of the most severe complications after HTx (2). To avoid the serious consequences of acute rejection (AR), immunosuppressants are extensively used to suppress allograft rejection and decrease the HTx death rate. However, the complications caused by immunosuppressant use, including infection, toxic reactions, and malignancy, could reduce the survival time of HTx patients (3). Therefore, it is urgent to find an appropriate substitute therapy to improve the long-term outcomes for HTx recipients.
Mesenchymal stem cells (MSCs), which are primarily derived from bone marrow and adipose tissue, play an important role in medicinal therapies owing to their ability to regulate the immune response (4). MSCs, which present homogeneous fusiform fibroblast-like morphology microscopically, express specific stem cell biomarker (CD44, CD73, CD90 >95%) and negatively express hematopoietic cell biomarker (CD45, CD34, CD11b <2%) which could be identified by flow cytometry (5). Besides their self-replicative and multidirectional differentiation potential, MSCs also have low immunogenicity and immunomodulatory properties, as confirmed by multiple animal studies and clinical trials (6,7). In addition, MSCs have been shown to induce immune tolerance and regulate the function of various immune cells, particularly macrophages, which support their potential use for treating rejection after heterogenous allograft organ transplantation (8-10).
Macrophages, which represent the major infiltrating immunocytes from peripheral blood circulation during the acute phase after HTx (11), are the chief antigen presenting cells participating in innate immunity (12). Macrophages can be polarized to M1 (classically activated macrophages) and M2 (alternatively activated macrophages) depending on the activation pathway and mode of immune function (13). The anti-inflammatory macrophages (M2 macrophages) characterized as overexpressing CD206 and Arg-1, have been found to substantially inhibit AR following HTx (14). Moreover, the M2 phenotype exhibits increased interleukin-10 (IL-10, anti-inflammatory cytokine) production (15). Indeed, MSCs can ameliorate immune rejection via promotion of macrophages toward the M2 phenotype (16). Nevertheless, the underlining mechanisms behind MSCs and macrophage polarization remains undetermined. Hence, enhancement of infiltrating MSCs in transplanted heart tissues and promotion of M2 polarization are considered critical aspects for reducing the occurrence of AR.
It is well known that the C-X-C motif chemokine receptor 4 (CXCR4) is the specific receptor of stromal cell-derived factor-1 (SDF-1) and this interaction is crucial for mediating the migration of tumor cells as well as stem cells (17,18). Moreover, overexpression of CXCR4 on the surfaces of MSCs has demonstrated therapeutic potential in cardiovascular diseases (19). Additionally, surface expression of CXCR4 on tumor cells reportedly boosts macrophage polarization to the M2 phenotype within the tumor microenvironment, ultimately aggravating tumor progression (20). However, it has not yet been reported whether surface expression of CXCR4 by MSCs similarly influences macrophage polarization. Therefore, we were aimed to demonstrate the role of CXCR4 on functions of MSCs, especially on the function of macrophage polarization, thereby potentially reducing the severity of AR after HTx.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs of 20–22 nucleotides that regulate various cell processes by affecting various of genes expression at the post-transcriptional level (21). Specifically, they bind to the 3'-untranslated region (3'-UTR) of target messenger RNAs (mRNAs) to promote mRNA degradation and inhibit translation (22). They can also regulate and control the differentiation and immunomodulatory properties of MSCs (23). The miRNA-204-3p has been reported to prevent the proliferation of bladder tumor cells by regulating lactate dehydrogenase-mediated glycolysis (24). It has also been confirmed recently that miRNA-204-3p mediated memory deficits in mouse model of Alzheimer’s disease by targeting Nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 4 (Nox4) (25). Nevertheless, it has not been studied in MSCs. Interestingly, we predicted miRNA-204-3p as the upstream miRNA of CXCR4. In the present study, we first demonstrated miRNA-204-3p/CXCR4 axis-mediated immunomodulatory functions of MSCs in acute rejection after heart transplantation. Thus, our findings may achieve the therapeutic goal of reduced AR following HTx. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-1293).
Methods
Isolation, in vitro culture, and labeling of MSCs
We extracted MSCs from the bone marrow of femurs and tibias of Wistar rats (male, 2-week-old) under general anesthesia using 1% pentobarbital sodium (40 mg/kg). The cell-culture medium used was Minimum Essential Medium-α (α-MEM, Biological Industries, Beit-Haemek, Israel) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Beit-Haemek, Israel). The bone marrow was flushed with culture medium and then cultured at 37 °C with 5% CO2. Cell purification was accomplished by washing with phosphate-buffered saline (PBS) to remove the non-adherent cells, and changing the culture medium after 48 h. The adherent cells were observed microscopically. Cells at passage 3 were utilized in the following experiments. The animal experiments were performed under the project license (SCXK Lu 20090001) granted by Shandong University Animal Research Ethics Committee, in compliance with institutional guidelines for the care and use of animals.
Cell-surface phenotype analysis of MSCs
The MSCs cultured at passage 3 showed a homogeneous fusiform fibroblast-like morphology. Flow cytometry was performed to authenticate the phenotype of the cultured cells. These MSCs were then incubated with the fluorescently labeled antibodies CD11b-PE, CD44-APC, CD45-FITC, and CD90-PE (Elabscience Bioscience, Wuhan, Hubei, China) using an AccuriC6 Plus flow cytometer [Becton, Dickinson and Co. Biosciences (BD), San Jose, CA, USA]. Antibodies of the relevant isotype were served as the negative control (NC).
Transfection of MSCs
For the transfection of CXCR4, the CXCR4 overexpression lentiviruses (oe-CXCR4) cloned into lentiviral vector LV5 and vector control (oe-NC) were synthesized by GenePharma (Shanghai, China). Lentiviruses encoding short-hairpin RNA (shRNA) for CXCR4, and the NC shRNA both cloned into lentiviral vector LV3 were constructed by GenePharma (Shanghai, China). After transfection for 8–12 h, fresh medium was added. Cells were collected for quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blotting analysis 48 h after transfection.
For miR-204-3p transfection, MSCs were transfected with miR-204-3p mimics, NC mimics, miR-204-3p inhibitor, and NC inhibitor. All synthesized RNA were purchased from GenePharma (Shanghai, China). Transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol. Cells were harvested 24 h after transfection and the efficiency of transfection was detected by qRT-PCR and western blotting. The sequences of synthesized RNAs were as follows: shRNA-CXCR4, 5'- GGATAACTACTCCGAAGAAGT-3'; miR-204-3p mimics, 5'-GCTGGGAAGGCAAAGGGACGTT-3'; miR-204-3p inhibitor, 5'-AACGTCCCTTTGCCTTCCCAGC-3'.
MSC and macrophage co-cultures
Rat macrophages (RMa-bm, R1920) purchased from Zhong Qiao Xin Zhou Biotechnology (Shanghai, China) were seeded into a 6-well plate and incubated at a density of 3×105/well for 24 h, after which, the culture medium was replaced with Dulbecco’s Modified Eagle’s Medium (DMEM; Biological Industries, Beit-Haemek, Israel) containing 10% FBS. Subsequently, the transfected MSCs were co-cultured with macrophages at a 1:1 ratio in a transwell system (Corning, Inc., New York, NY, USA) for an additional 48 h. The suspension and macrophages were harvested for the following experiments.
RNA extraction and qRT-PCR
Total RNA was extracted using the miRcute miRNA isolation kit (Tiangen Biotech, Beijing, China), according to the manufacturer’s instructions. The concentration of total RNA was measured using a NanoDrop spectrophotometer. Complementary DNA (cDNA) was synthesized using the PrimeScript Reverse Transcriptase Reagent Kit (Accurate Biotechnology, Changsha, Hunan, China), according to the manufacturer’s instructions, thereafter, quantitative fluorescence PCR was performed to detect the expression level using the SYBR Premix Ex Taq Kit (Accurate Biotechnology, China), according to the manufacturer’s protocols. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA were used as an internal control. All the primers (Table 1) were designed and synthesized by GenePharma (Shanghai, China).
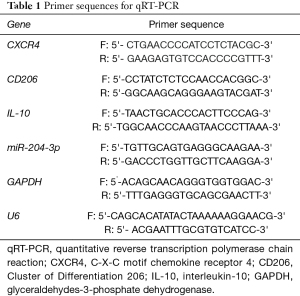
Full table
Western blot
Cells were washed thrice with cold PBS and lysed on ice in radioimmunoprecipitation assay (RIPA) buffer supplemented with 1% Phosphatase Inhibitor Cocktail III (MedChem Express, Monmouth Junction, NJ, USA). Protein lysates were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein samples were then transferred onto polyvinylidene fluoride (PVDF) membranes. Each membrane was washed in tris-buffered saline with Tween 20 (TBST) thrice, blocked with 5% bovine serum albumin (BSA) blocking-buffer (Beyotime Biotechnology, Shanghai, China) for 1 h at room temperature, and incubated at 4 °C overnight with one of the following rabbit anti-rat primary antibodies: anti-CXCR4 (1:800; Abcam, Cambridge, UK); anti-Arg-1 (1:1,000, Abcam, UK); anti-CD206 (1:500, Boster Biological Technology Company, Wuhan, Hubei, China), or anti-GAPDH (1:1,000; Abcam, UK). This was followed by incubation with horseradish peroxidase (HRP)-conjugated affinity goat anti-rabbit secondary antibody (1:5,000 MultiScience, Hangzhou, Zhejiang, China) for 1 h at room temperature. Immunoreactivity was detected using chemiluminescence reagents (Boster Biological Technology Company, Wuhan, Hubei, China).
Cell proliferation assay
Cell viability was assessed using the Cell Counting Kit-8 assay (CCK-8; MedChem Express, USA), according to the manufacturer’s protocol. Briefly, MSCs were seeded into 96-well plates at a density of 6×103 cells/well. Following overnight incubation, cell proliferation assays was performed on days 1, 2, 3, and 4, wherein a 10% CCK-8 solution was added to each well. The absorbance of the supernatant was measured at a wavelength of 450 nm using a microplate spectrophotometer (Thermo Fisher Scientific Inc., USA) after treating the cells with CCK-8 reagents for 2 h at 37 °C.
The 5-Ethynyl-2’-deoxyuridine (EdU) DNA Cell Proliferation Kit (Beyotime Biotechnology, China) was used according to the manufacturer’s instructions. The nuclei of proliferating cells appeared red under microscopy (×400).
The following formula was used:
Proliferation rate = number of proliferating cells (red)/total number of cells (purple) ×100%
Transwell migration assay
Cell migration was evaluated using transwell chambers (Corning Inc., USA) containing polycarbonate membrane filters (8 µm pore size). Preconditioned MSCs (1.5×105 cells/mL) in 200 µL serum-free medium were seeded in each upper chamber and 600 µL α-MEM with 10% FBS was placed into each lower well to induce cell migration. Following a 12 h incubation, cells on the upper side of the membrane were removed using cotton swabs, while cells on the bottom surface were stained with 0.1% crystal violet and fixed with 4% paraformaldehyde for 30 min. The cells on each chamber were then counted in randomly selected fields (×100).
Data acquisition, plasmid construction, and dual luciferase reporter assay
The upstream targets of CXCR4 were predicted using the TargetScan (www.targetscan.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), and miRBD (http://www.mirdb.org/miRDB/policy.html) databases. The wild-type and mutant CXCR4 3'-UTR sequences, designed and synthesized by GenePharma Co., Ltd. (Shanghai, China), were inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA), and both the wild-type and mutant luciferase reporter plasmids were constructed. The luciferase plasmids were then used to transfect 293T cells. Diluted plasmid DNA was mixed with miR-204-3p mimics and NC with the transfection reagent Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., USA) at room temperature for 20 min, and subsequently added to the culture wells. Following transfection for 48 h, fluorescence microscopy was used to detect the transfection efficiency of 293T cells, and green fluorescence (expression of green fluorescent protein, GFP) was considered successful. Luciferase activity at 560 nm was further determined by Dual-luciferase Reporter Gene System after transfection (Promega, E1910). By the way, the construction of luciferase reporter gene system to detect the interaction between miRNA-204 and CXCR4 was shown in Table 2.
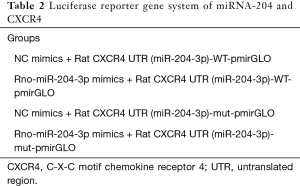
Full table
Enzyme linked immunosorbent assay for IL-10
To measure the concentration of IL-10, the supernatant collected from each well was centrifuged and subjected to a commercial enzyme-linked immunosorbent assay (ELISA) kit (Bioss, Beijing, China), according to the manufacturer’s instructions.
Statistical analysis
All experiments were independently repeated 3 times. All values are presented as the mean ± standard deviation. Data were analyzed using one-way analysis of variance (ANOVA) followed by Student’s t-tests (2-sided). A P value <0.05 was considered statistically significant. All analyses were performed using the SPSS 21.0 software (SPSS Inc., Chicago, IL, USA).
Results
Characterization of cultured MSCs
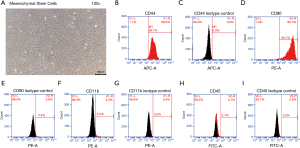
In vitro, MSCs cultured at passage 3 showed a homogeneous fusiform fibroblast-like form (Figure 1A). Flow cytometry demonstrated that the cells were uniformly negative for the hematopoietic markers CD11b and CD45, and positive for the stem cell antigens CD44 and CD90 (Figure 1B-1I). Thus, the phenotype of the cell population used in our study was consistent with that of MSCs.
CXCR4 promotes proliferation and migration of MSCs
To assess the effect of CXCR4 expression on MSC migration and proliferation, overexpression of CXCR4 cDNA (oe-CXCR4) and shRNA (sh-CXCR4) were introduced into rat MSCs through lentiviral infection. The transfection efficiency of CXCR4 was then analyzed by qRT-PCR and western blotting. In cells transfected with sh-CXCR4, CXCR4 expression was markedly deceased compared with that of NC (P<0.001). In contrast, cells transfected with oe-CXCR4 expressed significantly higher levels of CXCR4 compared with that of the NC (P<0.001; Figure 2A). The results were confirmed by western blotting (Figure 2B).
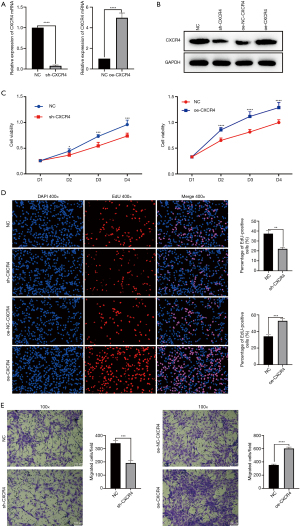
The proliferative effect of CXCR4 on MSCs was assessed by CCK-8 and EdU cell proliferation assays. Cells transfected with sh-CXCR4 exhibited reduced proliferation compared with that of NC, while cells overexpressing CXCR4 exhibited a higher level of active proliferation compared with that of the NC (Figure 2C,2D). The transwell migration assay also revealed that cells transfected with sh-CXCR4 had reduced migratory capacity compared with that of the NC, while cells overexpressing CXCR4 exhibited enhanced migration compared with that of the NC (Figure 2E).
Collectively, these results confirm the critical role of CXCR4 in MSC proliferation and migration, suggesting upregulation of CXCR4 may result in more viable MSCs being recruited into the myocardium.
CXCR4 of MSCs promote M2 macrophage polarization
To evaluate the effect that CXCR4-expressing MSCs have on macrophage polarization, MSCs were co-cultured with macrophages, and the supernatants were collected for analysis. The qRT-PCR revealed downregulated level of IL-10 and CD206 mRNA in macrophages co-cultured with sh-CXCR4-transfected MSCs, compared with that of the NC. Meanwhile, IL-10 and CD206 mRNA level was significantly upregulated in macrophages co-cultured with CXCR4-overexpressing MSCs (Figure 3A).
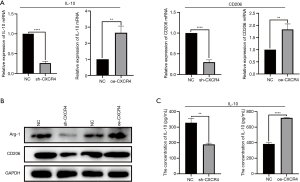
Furthermore, western blot analysis revealed that the abundance of Arg-1 and CD206 was markedly reduced in macrophages co-cultured with sh-CXCR4-transfected MSCs, compared with that of the NC. Alternatively, Arg-1 and CD206 protein abundance was upregulated in macrophages co-cultured with CXCR4-overexpressing MSCs (Figure 3B). These results related to IL-10 were further confirmed by ELISA (Figure 3C).
Taken together, these results suggest that upregulation of CXCR4 expression on MSCs supports the polarization of macrophages toward the M2 phenotype.
CXCR4 is a miR-204-3p target gene
After constructing a corresponding Venn diagram from the 3 databases, we identified the intersecting node (Figure 4A), from which miR-204-3p was selected as the candidate and the AAGGGUC sequence was predicted to be the binding site of miR-204-3p to the 3'-UTR of CXCR4 (Figure 4B), as demonstrated by dual luciferase reporter assay (Figure 4C).
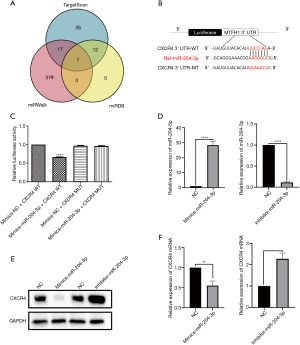
The MSCs were transfected with miRNA-204-3p mimics or its NC, or with miRNA-204-3p inhibitor or its NC. The qRT-PCR results showed that the expression of miRNA-204-3p in the mimics model was enhanced compared with that of the NC, while the expression of miRNA-204-3p in the inhibitor model was downregulated compared with that of the NC (Figure 4D).
Moreover, western blot analysis revealed that the abundance of CXCR4 protein was significantly downregulated in MSCs transfected with the miRNA-204-3p mimics compared with that of the NC, while its abundance was markedly upregulated in MSCs transfected with the miRNA-204-3p inhibitor compared with that of the NC (Figure 4E). These results were confirmed by qRT-PCR analysis (Figure 4F).
Anti-miR-204-3p promotes the proliferation and migration of MSCs by targeting CXCR4
To evaluate the effect of miR-204-3p on the proliferation of MSCs using CCK-8 assay, MSCs were transfected with miRNA-204-3p mimics or its NC; additionally, we co-transfected miRNA-204-3p mimics and the CXCR4 cDNA. The results showed that miRNA-204-3p mimics inhibited the proliferation of MSCs compared with that of the NC; whereas MSCs co-transfected miRNA-204-3p mimics and CXCR4 cDNA manifested better proliferative ability compared with cells transfected with miRNA-204-3p mimics.
Next, MSCs were transfected with miRNA-204-3p inhibitor or its NC; additionally, MSCs were co-transfected with miRNA-204-3p inhibitor and sh-CXCR4. The results showed that the miRNA-204-3p inhibitor promoted the proliferation of MSCs compared with that of the NC; whereas co-transfecting miRNA-204-3p inhibitor and sh-CXCR4 in MSCs partly weakened their proliferative ability compared with transfecting miRNA-204-3p inhibitor in MSCs (Figure 5A).
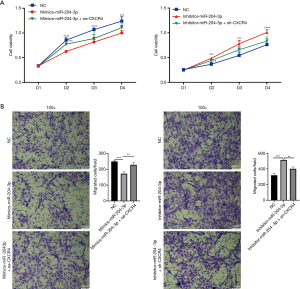
Similarly, to estimate the effect of miR-204-3p on MSC migration, transwell assays were performed. The results showed that miRNA-204-3p mimics inhibited the migration of MSCs compared with that of the NC; whereas MSCs co-transfected miRNA-204-3p mimics and oe-CXCR4 showed advantages in migratory capacity compared with MSCs transfected with miRNA-204-3p mimics. In addition, miRNA-204-3p inhibitor boosted the migratory activity of MSCs compared with that of the NC; meanwhile, co-transfecting miRNA-204-3p inhibitor and sh-CXCR4 in MSCs could partly weaken the migratory ability of MSCs caused by miR-204-3p inhibitor (Figure 5B).
Collectively, these results suggest that anti-miR-204-3p improved the number and migratory capacity of MSCs by targeting CXCR4.
Anti-miR-204-3p of MSCs promotes M2 macrophage polarization
To estimate the polarization effect caused by miR-204-3p, we co-cultured macrophages with MSCs that were transfected, or co-transfected, with oligonucleotides in the same manner as that described for Figure 5. The results revealed that the relative level of IL-10 and CD206 mRNA was downregulated in macrophages co-cultured with MSCs transfected with miRNA-204-3p mimics; whereas the miRNA-204-3p mimics + oe-CXCR4 group offset this effect. Meanwhile, the relative level of IL-10 and CD206 mRNA was upregulated in macrophages co-cultured with MSCs transfected with the miRNA-204-3p inhibitor compared with that of the NC; however, co-transfecting miRNA-204-3p inhibitor and sh-CXCR4 in MSCs partially dampened the relative expression level of IL-10 and CD206 mRNA in macrophages compared with transfecting miRNA-204-3p inhibitor (Figure 6A).
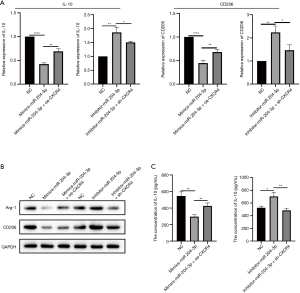
We also quantified the abundance of Arg-1 and CD206 protein and found that it was reduced in macrophages co-cultured with MSCs transfected with miRNA-204-3p mimics compared with that of the NC; however, MSCs co-transfected with miRNA-204-3p mimics and oe-CXCR4 promoted more Arg-1 and CD206 protein production in macrophages compared with MSCs transfected with miRNA-204-3p mimics. Meanwhile, the protein expression level of Arg-1 and CD206 increased in macrophages co-cultured with MSCs transfected with the miRNA-204-3p inhibitor compared with that of the NC; however, MSCs co-transfected miRNA-204-3p inhibitor and sh-CXCR4 partially lowered the expression of Arg-1 and CD206 in macrophages compared with MSCs transfected with miRNA-204-3p inhibitor (Figure 6B).
Finally, we found that IL-10 secretion by macrophages was reduced following co-culture with MSCs transfected with miRNA-204-3p mimics compared with that of the NC; however, MSCs co-transfected with miRNA-204-3p mimics and oe-CXCR4 partially elevated the concentration of IL-10 compared with MSCs transfected with miRNA-204-3p mimics. Alternatively, the concentration of secreted IL-10 increased following co-culture of macrophages with MSCs transfected with miRNA-204-3p inhibitor; however, MSCs co-transfected with miRNA-204-3p inhibitor and sh-CXCR4 degrade the concentration of IL-10 compared with MSCs transfected with miRNA-204-3p inhibitor (Figure 6C).
In summary, anti-miR-204-3p of MSCs promoted macrophage polarization toward the M2 phenotype via overexpression of CXCR4, while MSCs overexpressing anti-miR-204-3p may positively participate in immunosuppression during AR following HTx.
Discussion
As the incidence rate of cardiovascular diseases increases and clinical management improves, particularly that related to HF resulting from coronary heart disease in adults (26), and advancements in perioperative management are achieved, an increasing number of patients will benefit from HTx (1). Nevertheless, AR continues to negatively impact the HTx survival rate. Although this is ameliorated by immunosuppressant use, its disadvantages, such as the associated malignancy risk, are gradually gaining increasing attention (27).
Compared with chronic rejection, the seriousness of the threat of AR has been underestimated (28). Undeniably, AR remains the major cause of failure and even death of transplanted organs (29). In contrast to immunosuppressants, such as cyclosporin A, cell therapies based on MSCs present advantages not only in suppressing the functions of various immunocytes, but also in avoiding the severe adverse reactions (10). As far as the performance of MSCs recruited into the myocardium is concerned, it is imperative to ensure an increase in the number of MSCs via increased proliferation and migratory capacity.
The receptor for SDF-1a, CXCR4, exhibits strong chemotactic capacity and is thus regarded beneficial for promoting the infiltration of MSCs into injured heart tissues (30). In addition, our previous research has demonstrated that CXCR4 promotes survival and migration of MSCs under oxidative stress (31). Meanwhile, in the current study, we verified that CXCR4 overexpression promotes the proliferation and migration of MSCs. That is, overexpressing CXCR4 results in saturation of the transplanted heart with as many MSCs as possible. Consequently, we can achieve this goal by targeting CXCR4.
Additionally, we identified miR-204-3p as the target miRNA of CXCR4 as it exhibited the highest candidate score (96 points) in the miRDB database. We subsequently confirmed the relationship between miR-204-3p and CXCR4 using a dual luciferase reporter assay, which, to the best of our knowledge, represents the first report of this specific interaction. Furthermore, to investigate the effects of miR-204-3p, we constructed miR-204-3p mimics and inhibitor models and found that anti-miR-204-3p promoted both the proliferation and migration of MSCs. That is, anti-miR-204-3p effectively increased the number of MSCs to resist AR after HTx. Hence, this strategy has the capacity to potentially reduce the required drugs or even provide a substitute for them.
In the case of AR, accounting for the most frequent early postoperative complication of HTx, both cellular immunity and antibody-mediated rejection are involved (32). Microscopically, AR after HTx is characterized by varying degrees of intravascular and perivascular inflammatory cell infiltration of T cells and mononuclear macrophages accompanied by myocardial damage (32). Indeed, it is estimated that macrophages account for 38–60% of inflammatory cells in the heart, and other organs, during graft AR (33).
As the primary participating cell type in AR after allotransplantation, macrophages, which positively participate in both humoral and cellular immunity, promote inflammation by secreting various cytokines, such as IL-1, IL-6, tumor necrosis factor-α (TNF-α), and interferon-γ (INF-γ), or by participating in antigen presentation (34). After suffering AR, macrophages emerge from the peripheral circulation and swarm the graft in an effort to respond to donor antigens, consequently causing persistent myocytolysis, myocardial edema, and necrosis of myocardial cells (35). Moreover, macrophages boost AR by activating T cells and inducing the differentiation of T helper 17 (Th17) cells, which play vital roles in AR (36). Interestingly, macrophage polarization and biological function are determined by the local microenvironment. For instance, macrophages transform to the M2 phenotype (anti-inflammatory cells) expressing Arg-1 and CD206, and secreting the anti-inflammatory factor IL-10 when stimulated by IL-4 or IL-13 (13). Furthermore, M2 macrophages actively contribute to the inhibition of T-cell biological functions, which represent another significant contributor to AR after HTx (37). Thus, M2 macrophage polarization not only inhibits, or reverses, the proinflammatory function of macrophages but also interferes with the activity of other immune cells by severing the crosstalk between cells in the immune microenvironment. Moreover, MSCs can induce the polarization of M2 macrophages (38), thereby demonstrating their superior immunosuppressive effects. Thus, the underlining mechanism responsible for this effect of MSCs on macrophages is of great significance in guiding clinical treatment.
Additionally, overexpression of CXCR4 by tumor cells has been shown to enhance polarization of macrophages toward the M2 phenotype (tumor associated macrophage TAM), which in turn enhance the proliferation and metastasis of malignant cells (20,39). However, until now, it had remained unclear whether expression of CXCR4 by MSCs induces macrophages toward the M2 phenotype. In this study, we demonstrated that upregulation of CXCR4 caused an increase in the expression and secretion of IL-10 as well as the M2 macrophage specific markers CD206 and Arg-1. Collectively, these results indicated that CXCR4 promoted the polarization of macrophages from the M0 to M2 phenotype. Moreover, a positive association was detected between CXCR4 expression on MSCs and IL-10 concentration, suggesting an immunosuppressive role for CXCR4. These results further confirm the vital role that CXCR4 expressed on MSCs has in promoting the polarization of M2 macrophages.
Finally, we investigated the effect that miRNA-204-3p has on macrophage polarization. Collectively, our results demonstrated that miRNA-204-3p inhibits the polarizing effect of MSCs on macrophages, while overexpression of CXCR4 offsets this process. Moreover, transfection of MSCs with miRNA-204-3p inhibitor caused an increase in the number of M2 macrophages, whereas the anti-inflammatory activity was weakened following downregulation of CXCR4. Therefore, treatment of MSCs with anti-miRNA-204-3p was conducive to enhancing the polarization of macrophages toward the anti-inflammatory M2 phenotype, which could effectively enhance immunosuppressive effects following HTx.
Conclusions
In summary, surface expression of CXCR4 on MSCs not only promotes the proliferation and migration of MSCs, but also polarizing macrophages toward the anti-inflammatory M2 phenotype. Moreover, as the target miRNA of CXCR4, inhibition of miRNA-204-3p may effectively increase the number of MSCs infiltrating grafts and ameliorate AR following HTx by enhancing the proliferative capacity of MSCs and accelerating their migration. Furthermore, MSCs transfected with anti-miRNA-204-3p could contribute to the transformation of macrophages from the M0 phenotype to the M2 phenotype by targeting CXCR4. In summary, this study offered a new insight for the treatment of AR after HTx.
Acknowledgments
The authors thank the Key Laboratory of Cardiovascular Remodeling and Function Research, Qilu Hospital of Shandong University, Jinan, China, for their technical support.
Funding: This work was supported by the National Natural Science Foundation of China [81873510]; the Key Research Project of Shandong Province [2017GSF18146]; and the Youth Foundation of Qilu Hospital of Shandong University [2019QLQN21].
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-1293
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-1293
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-1293). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiments were performed under the project license (SCXK Lu 20090001) granted by Shandong University Animal Research Ethics Committee, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kim IC, Youn JC, Kobashigawa JA. The Past, Present and Future of Heart Transplantation. Korean Circ J 2018;48:565-90. [Crossref] [PubMed]
- Rivinius R, Helmschrott M, Rahm AK, et al. Risk factors and survival of patients with permanent pacemaker implantation after heart transplantation. J Thorac Dis 2019;11:5440-52. [Crossref] [PubMed]
- Bhat M, Mara K, Dierkhising R, et al. Immunosuppression, Race, and Donor-Related Risk Factors Affect De novo Cancer Incidence Across Solid Organ Transplant Recipients. Mayo Clin Proc 2018;93:1236-46. [Crossref] [PubMed]
- Cornelissen AS, Maijenburg MW, Nolte MA, et al. Organ-specific migration of mesenchymal stromal cells: Who, when, where and why? Immunol Lett 2015;168:159-69. [Crossref] [PubMed]
- Burnham AJ, Foppiani EM, Horwitz EM. Key Metabolic Pathways in MSC-Mediated Immunomodulation: Implications for the Prophylaxis and Treatment of Graft Versus Host Disease. Front Immunol 2020;11:609277 [Crossref] [PubMed]
- Shi Y, Xie J, Yang M, et al. Transplantation of umbilical cord mesenchymal stem cells into mice with focal segmental glomerulosclerosis delayed disease manifestation. Ann Transl Med 2019;7:383. [Crossref] [PubMed]
- Squillaro T, Peluso G, Galderisi U. Clinical Trials With Mesenchymal Stem Cells: An Update. Cell Transplant 2016;25:829-48. [Crossref] [PubMed]
- Wu J, Ji C, Cao F, et al. Bone marrow mesenchymal stem cells inhibit dendritic cells differentiation and maturation by microRNA-23b. Biosci Rep 2017;37:BSR20160436 [Crossref] [PubMed]
- Reinders ME, Hoogduijn MJ NK. Cells and MSCs: Possible Implications for MSC Therapy in Renal Transplantation. J Stem Cell Res Ther 2014;4:1000166 [Crossref] [PubMed]
- François M, Romieu-Mourez R, Li M, et al. Human MSC suppression correlates with cytokine induction of indoleamine 2,3-dioxygenase and bystander M2 macrophage differentiation. Mol Ther 2012;20:187-95. [Crossref] [PubMed]
- Wu G, Korsgren O, van Rooijen N, et al. The effect of macrophage depletion on delayed xenograft rejection: studies in the guinea pig-to-C6-deficient rat heart transplantation model. Xenotransplantation 1999;6:262-70. [Crossref] [PubMed]
- Kugelberg E. Macrophages: Controlling innate immune memory. Nat Rev Immunol 2015;15:596. [Crossref] [PubMed]
- Williams JW, Giannarelli C, Rahman A, et al. Macrophage Biology, Classification, and Phenotype in Cardiovascular Disease: JACC Macrophage in CVD Series (Part 1). J Am Coll Cardiol 2018;72:2166-80. [Crossref] [PubMed]
- Gao S, Mao F, Zhang B, et al. Mouse bone marrow-derived mesenchymal stem cells induce macrophage M2 polarization through the nuclear factor-κB and signal transducer and activator of transcription 3 pathways. Exp Biol Med (Maywood) 2014;239:366-75. [Crossref] [PubMed]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005;5:953-64. [Crossref] [PubMed]
- Melief SM, Schrama E, Brugman MH, et al. Multipotent stromal cells induce human regulatory T cells through a novel pathway involving skewing of monocytes toward anti-inflammatory macrophages. Stem Cells 2013;31:1980-91. [Crossref] [PubMed]
- Smit MJ, Schlecht-Louf G, Neves M, et al. The CXCL12/CXCR4/ACKR3 Axis in the Tumor Microenvironment: Signaling, Crosstalk, and Therapeutic Targeting. Annu Rev Pharmacol Toxicol 2021;61:541-63. [Crossref] [PubMed]
- Won YW, Patel AN, Bull DA. Cell surface engineering to enhance mesenchymal stem cell migration toward an SDF-1 gradient. Biomaterials 2014;35:5627-35. [Crossref] [PubMed]
- Zlabinger K, Lukovic D, Hemetsberger R, et al. Matrix Metalloproteinase-2 Impairs Homing of Intracoronary Delivered Mesenchymal Stem Cells in a Porcine Reperfused Myocardial Infarction: Comparison With Intramyocardial Cell Delivery. Front Bioeng Biotechnol 2018;6:35. [Crossref] [PubMed]
- Cai J, Zhang Q, Qian X, et al. Extracellular ubiquitin promotes hepatoma metastasis by mediating M2 macrophage polarization via the activation of the CXCR4/ERK signaling pathway. Ann Transl Med 2020;8:929. [Crossref] [PubMed]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015;16:421-33. [Crossref] [PubMed]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597-610. [Crossref] [PubMed]
- Xu C, Ren G, Cao G, et al. miR-155 regulates immune modulatory properties of mesenchymal stem cells by targeting TAK1-binding protein 2. J Biol Chem 2013;288:11074-9. [Crossref] [PubMed]
- Guo J, Zhao P, Liu Z, et al. MiR-204-3p Inhibited the Proliferation of Bladder Cancer Cells via Modulating Lactate Dehydrogenase-Mediated Glycolysis. Front Oncol 2019;9:1242. [Crossref] [PubMed]
- Tao W, Yu L, Shu S, et al. miR-204-3p/Nox4 Mediates Memory Deficits in a Mouse Model of Alzheimer's Disease. Mol Ther 2021;29:396-408. [Crossref] [PubMed]
- Lund LH, Edwards LB, Dipchand AI, et al. The Registry of the International Society for Heart and Lung Transplantation: Thirty-third Adult Heart Transplantation Report-2016; Focus Theme: Primary Diagnostic Indications for Transplant. J Heart Lung Transplant 2016;35:1158-69. [Crossref] [PubMed]
- Youn JC, Stehlik J, Wilk AR, et al. Temporal Trends of De Novo Malignancy Development After Heart Transplantation. J Am Coll Cardiol 2018;71:40-9. [Crossref] [PubMed]
- Mac QD, Mathews DV, Kahla JA, et al. Non-invasive early detection of acute transplant rejection via nanosensors of granzyme B activity. Nat Biomed Eng 2019;3:281-91. [Crossref] [PubMed]
- Berry GJ, Burke MM, Andersen C, et al. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J Heart Lung Transplant 2013;32:1147-62. [Crossref] [PubMed]
- Wu SZ, Li YL, Huang W, et al. Paracrine effect of CXCR4-overexpressing mesenchymal stem cells on ischemic heart injury. Cell Biochem Funct 2017;35:113-23. [Crossref] [PubMed]
- Bai X, Xi J, Bi Y, et al. TNF-α promotes survival and migration of MSCs under oxidative stress via NF-κB pathway to attenuate intimal hyperplasia in vein grafts. J Cell Mol Med 2017;21:2077-91. [Crossref] [PubMed]
- Gomaa AI, Ehsan NA, Elrefaei AA, et al. The Role of Monocyte/Macrophage and CXCR3 in Differentiation between Recurrent Hepatitis C and Acute Cellular Rejection Postliver Transplantation. J Immunol Res 2018;2018:2726939 [Crossref] [PubMed]
- Rowshani AT, Vereyken EJ. The role of macrophage lineage cells in kidney graft rejection and survival. Transplantation 2012;94:309-18. [Crossref] [PubMed]
- Ronca V, Wootton G, Milani C, et al. The Immunological Basis of Liver Allograft Rejection. Front Immunol 2020;11:2155. [Crossref] [PubMed]
- Gao C, Wang X, Lu J, et al. Mesenchymal stem cells transfected with sFgl2 inhibit the acute rejection of heart transplantation in mice by regulating macrophage activation. Stem Cell Res Ther 2020;11:241. [Crossref] [PubMed]
- Li B, Gurung P, Malireddi RK, et al. IL-10 engages macrophages to shift Th17 cytokine dependency and pathogenicity during T-cell-mediated colitis. Nat Commun 2015;6:6131. [Crossref] [PubMed]
- Oishi S, Takano R, Tamura S, et al. M2 polarization of murine peritoneal macrophages induces regulatory cytokine production and suppresses T-cell proliferation. Immunology 2016;149:320-8. [Crossref] [PubMed]
- Arabpour M, Saghazadeh A, Rezaei N. Anti-inflammatory and M2 macrophage polarization-promoting effect of mesenchymal stem cell-derived exosomes. Int Immunopharmacol 2021;97:107823 [Crossref] [PubMed]
- Wang D, Wang X, Si M, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 2020;474:36-52. [Crossref] [PubMed]


