
Relationship between HSPA1A-regulated gene expression and alternative splicing in mouse cardiomyocytes and cardiac hypertrophy
Introduction
Cardiac hypertrophy refers to an increase in the volume of cardiomyocytes and mainly occurs in situations of long-term pressure overload. Although the number of cardiomyocytes remains the same, the number of myocardial fibers increases, enhancing the contractility of the myocardium so that the heart can maintain normal blood circulation and reserve power. However, this compensatory function gradually develops into decompensation, resulting in myocardial ischemia, myocardial contractility decline, and eventuates in heart failure.
The main mechanisms involved in cardiac hypertrophy and cardiac remodeling are collagen synthesis and angiotensin II-induced cardiac fibroblast proliferation, increased myocardial oxygen consumption, and neuroendocrine activation, mediated by the sympathetic nervous system, renin angiotensin aldosterone system, vasopressin, and the interactions of multiple genes (1,2). Cardiac hypertrophy is categorized as pathological and physiological hypertrophy, which are associated with cardiac dysfunction and normal cardiac function, respectively. Pathological hypertrophy has a variety of causes including abnormal and prolonged hemodynamic tension due to myocardial infarction, and hypertension (3). Pathological hypertrophy is a complex process related to capillary rarefaction, fibrosis, increased production of pro-inflammatory cytokines, unfavorable epigenetic modifications, and cellular dysfunction (4). Emerging evidence has suggested an association between cardiac hypertrophy, the immune response, and activated inflammatory reactions (5-8). Cardiac hypertrophy eventuates in heart failure through the actions and remodeling effects of these factors. Many studies have confirmed that ventricular remodeling is the basic mechanism of heart failure, and cardiac hypertrophy in ventricular remodeling is an independent risk factor for cardiovascular disease. Therefore, reversing cardiac hypertrophy is of great significance in preventing the progression of heart failure and reducing mortality. With the overall increase in the incidence and mortality rate of heart failure, both the family and society such patients incur heavy burdens. Therefore, it is important to study the mechanisms of cardiac hypertrophy, including the factors involved in its development. Over the last decade, researchers have made significant progress in understanding cardiac hypertrophy and have identified numerous modifiers and signaling molecules associated with hypertrophy (9).
Hypertrophy and remodeling of the heart are pathological features of many heart diseases, and the underlying causes include hypertension, cardiomyopathy, valvular insufficiency, and myocardial infarction. In these diseases, ventricular hypertrophy occurs in response to pathological stimuli such as pressure and volume overload, sarcomere gene mutations, and neurohumoral activation. The main consequence of long-term and uncontrolled hypertrophic remodeling is cardiac insufficiency, which may lead to cardiac arrest due to heart failure or arrhythmias (10-12).
Previous studies have found that HSPA1A (HSP72/HSP70) is involved in the regulation of cell proliferation and apoptosis, which may be a potential therapeutic window for cardiac hypertrophy and other related diseases (13-17).
The HSPA1A gene is associated with the occurrence and development of a variety of diseases (18-21). Hsp70 is a strongly conserved 70 kDa heat shock protein; the gene is located in the same area as the histocompatibility complex III. The HSPA1A gene encodes an RNA-binding protein HSP70 which has been shown to influence the development of cardiac hypertrophy (22-24). Studies have shown that loss of HSP70 leads to cardiomyocyte dysfunction and an impaired ischemia/reperfusion stress response in the heart (25-27). In addition, deletion of the HSPA1A gene may lead to cardiac insufficiency and cardiac hypertrophy through activation of JNK, p38MAPK, RAF-1, and ERK (26-29).
Stimulation of hypertrophy can induce the expression of HSP70. Induced HSP70 is physically related to and activated by histone deacetylase 2 (HDAC2) (30-32). Myocardial hypertrophy and HDAC2 activation are significantly impaired in HSP70-deficient mice. Studies have shown that the induction of HSP70 under different hypertrophic stress responses and the activation of HDAC2 trigger cardiac hypertrophy, which suggests the hypertrophic regulatory mechanism of HSP70/HDAC2 (33-35). There is a study aimed to examine the influence of NF-κB p65 in chronic constriction injury (CCI)-induced neuropathic pain as well as its underlying mechanism.NF-κB p65 and HDAC2 were upregulated while miR-183 was downregulated in the dorsal horn of the CCI rat spinal cord. NF-κB p65 was bound to the HDAC2 promoter and then increased its expression. HDAC2 reduced miR-183 expression by deacetylation of histone H4. Additionally, miR-183 negatively regulated TXNIP. Mechanistically, NF-κB p65 downregulated the miR-183 expression via the upregulation of HDAC2 and further induced inflammatory response by activating the TXNIP-NLRP3 inflammasome axis, thus aggravating the neuropathic pain in CCI rats and microglial cells.
All this evidence indicates that HSP70 has gene regulatory functions. However, whether such functions are involved in cardiac hypertrophy, together with the mechanisms involved, is still unclear.
In this study, the relationship between HSPA1A (HSP72/HSP70) and the development of cardiac hypertrophy and its mechanism of action were studied from the perspective of RNA-binding proteins and their functions. The study of RNA-binding proteins is currently a hotspot in RNA research. In this study, we hypothesized that HSPA1A may regulate the expression of target genes by affecting the activity of transcription factors and may also regulate the expression levels of target genes, including immune- and inflammation-related genes, by binding RNA to modulate transcription factor alternative splicing (AS), ultimately influencing the development of cardiac hypertrophy.
In this study, we investigated non-coding (nc) RNAs and HSPA1A-regulated genes in HL-1 cells and, in addition, created a mouse model of cardiac hypertrophy, through transverse aortic constriction (TAC), performing high-throughput sequencing on ventricular tissue samples from the mice. Transcriptomic data was obtained by paired-end sequencing on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA, USA). We identified differentially expressed genes (DEGs) by analysis of data from the different experimental groups. We overexpressed the HSPA1A gene in mouse heart HL-1 cells which are widely used to study cardiac hypertrophy, and analyzed changes in transcriptome expression through high-throughput sequencing. In addition, we investigated inflammation- and immune-related genes potentially involved in cardiac hypertrophy, using quantitative reverse transcription polymerase chain reaction (RT-qPCR) for verification. Finally, to understand the behavior of HSPA1A in AS, all transcriptomic sequencing data were investigated for AS events using ABLas software. The overall aim to investigate the relationship between HSPA1A regulation of mouse cardiomyocyte gene expression and cardiac hypertrophy. Through this research, we hoped to further expand our understanding of the clinical treatment goals of cardiac hypertrophy, and provide clinical prevention and better treatment methods for cardiac hypertrophy-related heart failure.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-1222).
Methods
Ethical approval
All procedures were approved by the Institutional Animal Care and Use Committee of Jilin University and conducted in accordance with the Guideline for Ethical Review of Animal Welfare and Guide for the Care and Use of Laboratory Animals.
TAC
The surgical method of TAC is often used to investigate the progression from cardiac hypertrophy to heart failure owing to pressure overload within the hearts of mice. This induces afterload enhancement in the heart resulting in hypertrophic compensation over the following few weeks. The degree of constriction indicates the TAC severity and is determined by the gauge of the needle used. The operation usually adopts a midline incision of the neck and chest; after separating the muscle, midline thoracotomy, and stripping the thymus, the aortic arch is visualized and narrowing of the aortic arch is performed. The chest is then closed and the skin of the head and neck sutured. After the surgery, the mice generally show rapid recovery and less immune response to trauma. Pathological cardiac hypertrophy was established via TAC operation under isoflurane anesthesia (the mice were rendered unconscious after having inhaled an appropriate amount of isoflurane). All TAC mice with cardiac hypertrophy were compared with sham-operated mice (similar method without the tying).
Animal models
Healthy male (C57BL/6J) mice (8-week-old; body weight 23–33 g) were utilized in all investigations. All mice were divided into four groups: TAC + dantrolene, TAC + DMSO, sham + dantrolene, and sham + DMSO. The mice in each group were given drugs (dantrolene, 30 mg/kg, per day, for 4 weeks. The volume of DMSO was the same as that of dantrolene). Dantrolene inhibits the release of calcium from the sarcoplasmic reticulum by binding and blocking the ryanodine receptor, thus inhibiting muscle contraction and other calcium-dependent cytoplasmic metabolism processes, which may delay cardiac hypertrophy and ventricular remodeling and improve heart failure. After sacrifice at 4 weeks, the heart and tibia were collected to measure the dry weight of the heart and the length of both tibias, and the average value was taken to calculate the ratio of heart weight to tibia length and the ratio of heart weight to body weight. High-throughput sequencing was conducted on the four groups using the Illumina HiSeq X Ten platform. The DEGs were identified by differential expression analysis of the transcriptomic data. A protocol was prepared before the study without registration.
Cell culture and transfections
The HL-1 cardiac muscle cell line (Procell Life Science & Technology Co., Ltd., Wuhan, China) cells were cultured at 37 °C with 5% CO2 in Minimum Essential Media (MEM) with 10% fetal bovine serum (FBS), 100 µg/mL streptomycin, and 100 U/mL penicillin. According to the protocol of the manufacturer, plasmid transfection of HL-1 cells was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, the transfected cells were analyzed by RT-qPCR.
RNA extraction and sequencing
The HL-1 cells were homogenized before RNA extraction. We used RQ1 DNase (Promega, Madison, WI, USA) to remove DNA. Purified RNA was quantitated by measuring the 260 nm/280 nm absorbance in a Smart Spec Plus spectrophotometer (BioRad, Hercules, CA, USA) and its purity was confirmed by 1.5% agarose gel electrophoresis. For all specimens, 1 µg of total RNA was used to prepare the RNA sequence library using the Kapa chain mRNA sequence kit (kk8541) on the Illumina platform. Polyadenylated messenger RNAs (mRNAs) were purified and fragmented. The mRNA fragments were reverse-transcribed into double-stranded circular DNA (cDNA). After end and tail repair, the DNA was connected to the diluted Roche (Basel, Switzerland) adapter. After purification, the ligation products were divided into segments of 300–500 bp, amplified, purified, quantified, and stored at −80 °C before sequencing. The deoxyuridine triphosphate (dUTP)-labeled strand (the second cDNA strand) was not amplified and strand-specific sequencing was allowed. To achieve a high sequencing throughput, libraries were prepared according to the manufacturer’s instructions and applied to the Illumina Novaseq 6000 system for sequencing the 150 nt paired-end.
RNA-seq raw data cleaning and alignment
The original read involving more than a 2-N base was discarded. The FASTX Toolkit (Version 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/) was then utilized to cut out the adapter and low-quality bases from the original sequencing read. Further, short reads smaller than 16 NT were deleted. After that, clean reads were aligned to the GRch38 genome by tophat2 (34-41) allowing 4 mismatches. Unique mapping reads were used for gene reading counts and FPKM calculations (fragments per kilobase of the transcript per million fragments mapped).
DEG identification
The R Bioconductor edgeR package (https://bioconductor.org/packages/release/bioc/html/edgeR.html) was used to record the DEGs. An erroneous finding rate <0.05 and fold change >2 or <0.5 were utilized as the cut-off criteria.
AS measurement
The ABLas pipeline was used to define and quantify the regulated alternative splicing events (RASEs) and alternative splicing events (ASEs) between the samples. ABLas utilized 10 categories of ASEs regarding the splice junction reads, including alternative 5' splice site (A5SS), exon skipping (ES), intron retention (IR), mutually exclusive 5'UTRs (5pMXE), mutually exclusive exons (MXE), alternative 3' splice site (A3SS), mutually exclusive 3'UTRs (3pMXE), cassette exon, A3SS&ES, and A5SS&ES. To evaluate these, Student’s t-test was applied to determine the significance of event ratio change. Special events which were considerable at a P value cutoff conforming to an erroneous finding rate cutoff of 5% were taken into account as RBP-regulated ASEs.
qRT-PCR validation of DEGs and AS events
In the current research, qRT-PCR was carried out for some of the DEGs to verify the RNA-seq data. The remaining total RNA from the preparation of the RNA-seq library was used for RT-qPCR. The RNA was reverse-transcribed into cDNA by M-MLV Reverse Transcriptase (Vazyme Biotech Co., Ltd., Nanjing, China). The SYBR Green PCR Reagents Kit (Yeasen Biotechnology, Shanghai, China) was used for real-time PCR with the system of Step One Real-time PCR. The PCR conditions included denaturing for 10 min at 95 °C, 40 denaturing cycles for 15 s at 95 °C, and extension and annealing for 1 min at 60 °C. The PCR amplifications were performed in triplicate for each specimen. The RNA expression levels of all genes were standardized with that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and the ASE were verified by qRT-PCR. To detect alternative isoforms in the sequences, we used a boundary-spanning primer containing the junction of constitutive exon and alternative exon and the opposite primer in the fundamental exon. According to “model exon”, we designed the boundary-spanning primer of the alternative exon to detect model splicing or according to “altered exon” we designed the primer to detect altered splicing.
Functional enrichment measurement
To categorize the functional classifications of the DEGs, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms were determined by the KOBAS 2.0 server (http://kobas.cbi.pku.edu.cn/). The degree of enrichment of each term was determined by the hypergeometric method and Benjamini-Hochberg false discovery rate (FDR) monitoring program.
Availability of materials and data
The outcomes discussed in this paper can be obtained under the Gene Expression Omnibus (GEO) Series accession number GSE158635.
Cloning and plasmid construction
The design of primer pairs for Hot Fusion was carried out through CE Design V1.04. Each considered primer contained a segment of 17–30 bp sequence of the pIRES-hrGFP-1a vector as well as a gene-specific sequence.
F-primer: agcccgggcggatccgaattcATGGCCAAAGCCGCGGCG;
R-primer: gtcatccttgtagtcctcgagATCTACCTCCTCAATGGTGGGG.
The digestion of pIRES-hrGFP-1a vector was performed with XhoI (NEB, Ipswich, MA, USA) and EcoRI for 2–3 h at 37 °C. The results of the enzyme digestion were analyzed on 1.0% agarose gels and also refined using the Qiagen (Hilden, Germany) column kit. A TRIzol reagent was used for obtaining total RNA from the cells. The purified RNA was reverse-transcribed into cDNA using the oligodT primer and the inserted segment was amplified by PCR. A linearized vector digested by EcoRI and XhoI (NEB) and PCR insert were added to a PCR microtube for ligation with ClonExpress® II One Step Cloning Kit (Vazyme). The plasmids were transformed into Escherichia coli. The transformed cells were cultured on luria broth (LB) agar plates with 1 µL/mL ampicillin overnight at 37 °C. Colonies were monitored by colony PCR (28 cycles) with universal primers (placed on the backbone vector).
Measurement of gene overexpression
We used GAPDH as a control to evaluate the impact of HSPA1A overexpression. The cDNA was synthesized through the benchmark procedure and RT-qPCR was carried on the Bio-Rad S1000 with Bestar SYBR Green RT-PCR Master Mix (DBI Bioscience, Shanghai, China). Transcript concentrations were normalized to the GAPDH mRNA level by the 2−ΔΔCT method. The software GraphPad Prism (San Diego, CA, USA) was used to analyze resulted with paired Student’s t-tests.
Statistical analysis
The results are presented as mean ± standard deviation (SD). We used t-tests for paired samples and equal variance. Statistical significance was considered when P<0.05.
Results
Animal models and determination of HSPA1A
A pathological cardiac hypertrophy model was established by exposing C57BL/6J mice to pressure overload via TAC. It was found that the hearts were considerably expanded in all model mice as shown by the heart weight to tibia length (HW/TL) (P<0.001) and the heart weight to body weight (HW/BW) ratios (Figure 1A,1B).

The identified DEGs encoding RNA-binding proteins are shown in Tables 1-5. It can be seen that that HSPA1A was significantly differentially expressed in both the TAC + dantrolene vs. sham + dantrolene and the TAC + DMSO vs. sham + DMSO groups. The RNA-binding proteinZfp36 was also significantly differentially expressed between both the TAC + dantrolene vs. sham + dantrolene group and the TAC + DMSO vs. sham + DMSO group. These results suggest that Zfp36 is related to cardiac hypertrophy. The expression of MKI67 and GM5619 was significantly different between the TAC + dantrolene and TAC + DMSO groups. These results suggest that dantrolene may delay cardiac hypertrophy and ventricular remodeling and improve heart failure by regulating the expression of RNA binding protein genes MKI67 and GM5619. From the research results of dantrolene’s gene regulation, this drug may have the potential to become a drug for the treatment of myocardial hypertrophy.
Table 1
| N_Med_ |
Y_Med_ |
Y_NC_ |
Y_Med_ |
|---|---|---|---|
| Bclaf1 | Hspa1b | PCDHGA9 | MKI67 |
| Prkdc | Zfp36 | GSPT2 | Gm5619 |
| Lin28b | HSPA1A | INTS6 | |
| Rc3h2 | Khdrbs2 | SNRNP48 | |
| Mbnl3 | Myod1 | FMR1 | |
| Tfrc | MKI67 | ||
| Srek1 | ZCCHC7 | ||
| Khdrbs2 | CELF6 | ||
| Cdkn2a | RNPC3 | ||
| Mex3a | DNTTIP2 | ||
| CLK1 | |||
| ZFP36 | |||
| TOP1 | |||
| PPP1R10 | |||
| HSPA1B | |||
| PNISR | |||
| HSP90AA1 | |||
| EIF5 | |||
| HSPA1A |
DEGs, differentially expressed genes; Y, tac; N, sham; Med, dantrolene; NC, DMSO.
Table 2
| logFC | logCPM | P Value | FDR | Symbol | |
|---|---|---|---|---|---|
| ENSMUSG00000037608.13 | 1.37691317 | 3.298172462 | 0.004976583 | 0.272670487 | Bclaf1 |
| ENSMUSG00000022672.8 | 1.559831186 | 3.525988301 | 0.001517032 | 0.140376064 | Prkdc |
| ENSMUSG00000063804.7 | 1.537520665 | 1.521149498 | 0.002666848 | 0.191267309 | Lin28b |
| ENSMUSG00000075376.8 | 1.289934547 | 3.873883786 | 0.008061189 | 0.377370845 | Rc3h2 |
| ENSMUSG00000036109.15 | 1.674792316 | 2.460041165 | 0.000842666 | 0.096055768 | Mbnl3 |
| ENSMUSG00000022797.13 | -1.645728356 | 3.347068776 | 0.000826715 | 0.094811756 | Tfrc |
| ENSMUSG00000032621.7 | 2.342676789 | 2.822109246 | 4.48E-06 | 0.005686282 | Srek1 |
| ENSMUSG00000026058.9 | 1.408480248 | 3.498172685 | 0.004062046 | 0.243269108 | Khdrbs2 |
| ENSMUSG00000044303.6 | 1.449735526 | -0.022150109 | 0.009478601 | 0.414179768 | Cdkn2a |
| ENSMUSG00000074480.4 | 1.464034534 | 2.506879331 | 0.003275334 | 0.210130768 | Mex3a |
FC, fold change, expressing the absolute ratio of change; FDR, false discovery rate, expressing the statistical reliability of changes; Criterion of significant difference expression: FC ≥2 or ≤0.5, P value <0.01.
Table 3
| Gene | logFC | logCPM | P value | FDR | Symbol |
|---|---|---|---|---|---|
|
|
2.778137388 | 5.020628632 | 1.90E-05 | 0.030450357 | Hspa1b |
|
|
−2.115314018 | 7.286336698 | 0.000815024 | 0.264537571 | Zfp36 |
|
|
2.372585928 | 4.982064634 | 0.000204457 | 0.112807995 | HSPA1A |
|
|
−1.648019437 | 3.401815444 | 0.008649659 | 0.623748237 | Khdrbs2 |
|
|
−3.715601599 | −2.705172887 | 0.004270773 | 0.504099998 | Myod1 |
FC, fold change, expressing the absolute ratio of change; FDR, false discovery rate, expressing the statistical reliability of changes; Criterion of significant difference expression: FC ≥2 or ≤0.5, P value <0.01.
Table 4
| Gene | logFC | logCPM | P value | FDR | Symbol |
|---|---|---|---|---|---|
|
|
−2.806795858 | −2.325968383 | 0.003198221 | 0.178604228 | Pcdhga9 |
|
|
2.123629649 | −2.090441631 | 0.00885937 | 0.309251373 | Gspt2 |
|
|
1.488913794 | 0.748277562 | 0.005898055 | 0.242972883 | Ints6 |
|
|
1.664764747 | 0.415504795 | 0.002674302 | 0.161278361 | Snrnp48 |
|
|
1.583785592 | 0.613858941 | 0.00422027 | 0.206158886 | Fmr1 |
|
|
2.148112722 | −1.069390741 | 0.001258408 | 0.102956221 | Mki67 |
|
|
1.552881197 | −0.350847883 | 0.007382847 | 0.28140846 | Zcchc7 |
|
|
4.964133167 | −3.038231362 | 0.002089194 | 0.136710783 | Celf6 |
|
|
1.486210486 | 0.526115757 | 0.005861614 | 0.242394671 | Rnpc3 |
|
|
1.514988909 | 2.189779488 | 0.003584803 | 0.185758765 | Dnttip2 |
|
|
1.321303497 | 2.692015099 | 0.009882728 | 0.326418146 | Clk1 |
|
|
−2.266389273 | 7.078785436 | 1.28E-05 | 0.004035919 | Zfp36 |
|
|
1.346245201 | 2.127085147 | 0.009211278 | 0.312369117 | Top1 |
|
|
−1.608443229 | 4.932335719 | 0.0015964 | 0.120109846 | Ppp1r10 |
|
|
4.908482456 | 5.964209437 | 5.75E-17 | 1.35E-12 | Hspa1b |
|
|
1.454958988 | 1.287504692 | 0.006195431 | 0.252996303 | Pnisr |
|
|
2.143538118 | 4.845410675 | 3.56E-05 | 0.008678452 | Hsp90aa1 |
|
|
1.515316562 | 4.034093234 | 0.002968089 | 0.172333315 | Eif5 |
|
|
4.449406577 | 5.794099113 | 8.84E-15 | 5.17E-11 | HSPA1A |
FC, fold change, expressing the absolute ratio of change; FDR, false discovery rate, expressing the statistical reliability of changes; Criterion of significant difference expression, FC ≥2 or ≤0.5, P value <0.01.
Table 5
| Gene | logFC | logCPM | P value | FDR | Symbol |
|---|---|---|---|---|---|
|
|
−2.562908973 | −1.178255284 | 0.00104434 | 0.264217927 | Mki67 |
|
|
−5.068949218 | −3.125790227 | 0.003698223 | 0.500088957 | Gm5619 |
FC, fold change, expressing the absolute ratio of change; FDR, false discovery rate, expressing the statistical reliability of changes; Criterion of significant difference expression: FC ≥2 or ≤0.5, P value <0.01.
The HSPA1A-regulated transcriptome in HL-1 cells and 69 significantly upregulated genes and 52 downregulated genes
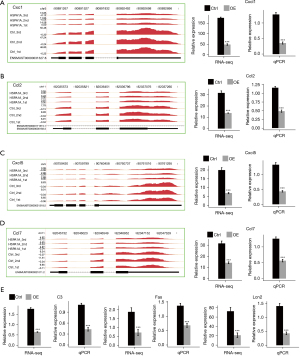
Overexpression (OE) of HSPA1A in mouse cardiac HL-1 cells (Figure 2A,2B) resulted in the differential expression of 121 DEGs, including 69 significantly upregulated genes and 52 downregulated genes [the cutoff was adjusted as fold change (FC) ≥2 or ≤0.5 and a 5% rate of erroneous finding (FDR)] (Figure 2C). The heatmap shows the DEG expression patterns were related to HSPA1A-mediated transcription (Figure 2D). The GO annotation and KEGG pathway analysis were used to analyze these 121 HSPA1A-regulated DEGs (Figure 2E-2H). In terms of the biological process GO analysis, the upregulated genes in HSPA1A-OE HL-1 cells were mainly enriched in the G protein-coupled receptor signaling pathway, signal transduction, and transportation processes. Notably, the expression of many ncRNAs was significantly upregulated in HSPS1A-OE HL-1 cells. The ncRNAs with the top expression levels, Rn7SK and Rmrp, have been reported to be associated with cardiac hypertrophy. The downregulated genes were mostly related to inflammatory and immune responses, including CXCL1, CCL2, CCL7, CXCL5, Fas, and c3. In addition, the highest expressed gene LCN2 among all downregulated genes is also an immune mediator and plays a key role in heart failure and the ontogeny of cardiac hypertrophy.
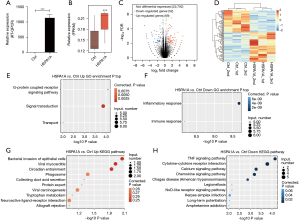
Verification of HSPA1A-regulated expression of ncRNAs and immune response-related genes associated with cardiac hypertrophy in HL-1 cells
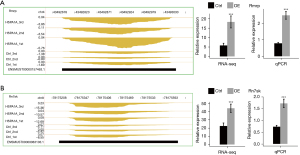
To confirm the expression of ncRNAs and genes regulated by HSPA1A in HL-1 cells, we performed RT-qPCR to verify the expression of the two highest expressed ncRNAs, Rn7sk and Rmrp, and 7 immune response-related genes, including CXCL1, CCL2, CCL7, CXCL5, Fas, C3, and LCN2. The PCR primer pairs (Table 6) were organized to specifically reinforce these ncRNAs and genes. The RT-qPCR verification results of ncRNA (Figure 3A,3B) and immune response-related genes (Figure 4A-4E) were all consistent with the RNA-seq results. The data indicated that HSPA1A positively regulates the expression of Rn7sk and Rmrp in HL-1 cells and negatively regulates the expression of genes associated with the immune and inflammatory responses associated with cardiac hypertrophy.
Table 6
| Primer | Sequence (5' to 3') | |
|---|---|---|
| 01 | Rn7sk-F | CCTACAATGGACCTTGAGAG |
| 02 | Rn7sk-R | TCGAAGAGGACGACCTTC |
| 03 | Rmrp-F | CTGCCTGCGTCACTATGT |
| 04 | Rmrp-R | CCGCAAGTCACTGTTAGC |
| 05 | Cxcl1-F | CTCTGATGGCACCTGTCT |
| 06 | Cxcl1-R | TGAACAAGCAGAACTGAACT |
| 07 | Lcn2-F | CAGATGCTCCTTGGTATGG |
| 08 | Lcn2-R | TGAATGGGTGGTGAGTGT |
| 09 | Ccl2-F | TGAGATCAGAACCTACAACT |
| 10 | Ccl2-R | TCCTACAGAAGTGCTTGAG |
| 11 | Ccl7-F | TGAAGCCCATCAGAAGTG |
| 12 | Ccl7-R | GAACAGCGGTGAGGAATT |
| 13 | Cxcl5-F | AAGAAGGTAAGAAGCAAGGA |
| 14 | Cxcl5-R | AAGACACACTACAGACATCT |
| 15 | Fas-F | ATGACTAGCAGGTTCCAG |
| 16 | Fas-R | GCAGATTCCACTTCACATT |
| 17 | C3-F | ACCACCATAGATTCTGTGAA |
| 18 | C3-R | GCTACATCATTGGGAAGGA |
HSPA1A regulates the AS of Asxl2 and Runx1
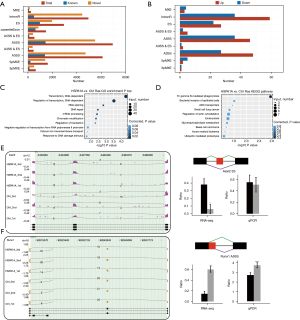
To investigate the effect of HSPA1A on AS, all ASEs were identified from the transcriptome sequencing data and further analyzed using the ABLas software tool. We identified known ASEs in the model gene and reference genome (Figure 5A). By exerting a rigorous cutoff of P value of ≤0.05 and modified AS ratio of ≥0.2, we determined 366 credible RASEs, A5SS events, ES events, A3SS events, and IR events (Figure 5B). This indicated that significant regulation of the genes via HSPA1A in alternative splicing levels (RASGs) were greatly enriched in transcription, regulatory transcription, RNA splicing, DNA repair, mRNA processing, chromatin modification, regulatory translation, negative transcription regulation of RNA polymerase II promoter, DNA damage stimulation response (GO biological process terms, Figure 5C), and calcium ion transmembrane transport. Moreover, KEGG analysis showed significant enrichment of the genes regulated via HSPA1A in AS in bacterial invasion of epithelial cells, Fc gamma R-mediated phagocytosis, ABC transporters, small cell lung cancer, regulation of actin cytoskeleton, glycerophospholipid metabolism endocytosis, basal cell carcinoma, acute myeloid leukemia, and ubiquitin-mediated proteolysis (Figure 5D). Among these RASGs related to the overexpression of HSPA1A in HL-1 cells, Asxl2 and Runx1 are associated with cardiac hypertrophy. The consistent RT-qPCR verification results of ASEs in Asxl2 and Runx1 (Figure 5E,5F), compared to the RNA-seq results suggest that HSPA1A can function in cardiac hypertrophy by regulating the AS of Asxl2 and Runx1.
Discussion
The HSPA1A gene encodes a heat shock protein involved in stress protection in cells. It has been reported to participate in cellular regulation of apoptosis and proliferation, as well as the activation of transcription factors and protein degradation. It has been reported that hypertrophic stimulation can induce Hsp70 expression, which consequently causes cardiac dysfunction and the development of cardiac hypertrophy by activating JNK, p38-MAPK, Raf-1, and ERK (41). Studies have also revealed Hsp70 induction under conditions of different hypertrophic stress responses and subsequent activation of HDAC2 to trigger cardiac hypertrophy (42). Nevertheless, the detailed mechanisms require further investigation.
In this study, we investigated the role of HSPA1A in cardiac hypertrophy and whether the gene can regulate gene expression and AS in HL-1 cells. We used analysis of both HSPA1A expression in pathological cardiac hypertrophy induced in an animal model and overexpression of the gene in mouse cardiac HL-1 cells.
Overexpression of HSPA1A in HL-1 cells resulted in the significant upregulation of 69 genes. Although these genes were not enriched in the functional pathways associated with cardiac hypertrophy, 2 highly expressed ncRNAs, Rn7sk and RMRP, were detected in the cardiomyocytes. It was also found that HSPA1A also negatively regulated the expression of 52 genes, which were enriched in both inflammation and the immune response, 2 biological processes related to cardiac hypertrophy. Further, the CXCL1, CCL2, CCL7, CXCL5, Fas, and C3 genes were principally related to cardiac hypertrophy. Additionally, LCN2, the topmost gene in the ranking is also associated with cardiac hypertrophy. In addition, 366 variable splicing events with considerable modifications were discovered after HSPA1A overexpression. The most relevant functional pathways corresponding to the abovementioned events are transcription and transcriptional regulation.
A study showed that cardiac hypertrophy in mice originated from the Dnmt2-deficiency. An essential level for cardiac improvement, namely, activation of the P-TEFb complex, stems from the enhanced separation of Rn7sk ncRNA component in Dnmt2-deficient cells which were negatively regulated (43,44). The reported results showed that among conserved ncRNAs, Rmrp, h19, and HOTAIR were also induced in a mouse model of heart hypertrophy (45). There is a study showing that angiotensin II-induced penetration of monocytes within the heart is strongly influenced by CXCL1–CXCR2 signaling which establishes and aggravates the remodeling of cardiac hypertrophy. Inactivation or inhibition of CXCL1 and/or CXCR2 could represent novel therapeutic objectives for the treatment of hypertensive heart maladies (46). An investigation has shown that the genetic elimination of monocyte chemoattractant protein (MCP)-1 (MCP-1-KO mice) hindered the Ang-II-induced cardiac fibrosis as well as the fibroblast expression of CD34+/CD45+ (47). There is data suggesting inhibition of CXCL2 and CXCL5 chemoattractant expression via 11β-HSD1 as a modern strategy with the potential for neutrophil regulation recruitment to the damaged myocardium, and fibroblasts of cardiac as the main site for the regeneration of intracellular glucocorticoid affected by severe inflammation after myocardial trauma (48). Also, there are reports that FasL and Fas are expressed in the cardiomyocytes and myocardium. Myocardial expression of Fas is almost associated with diastolic loading in vivo circumstances. Signaling initiated by Fas in cardiomyocytes is coupled to activation of the AP-1 transcription factor. The previous study results show that CC, through C3, participates in myocardial regeneration and preservation in response to acute myocardial infarction. This study showed that the direct impact of LCN2 on cardiomyocyte number and size, together with a consistent association of LCN2 in the ontogeny of heart failure and cardiac hypertrophy. Both Asxl2 and Runx1 are also associated with cardiac hypertrophy (43-48).
The results of the present study suggest that HSPA1A could act to induce cardiac hypertrophy by promoting ncRNA expression at the transcriptional level and inhibiting the inflammatory and immune responses in cardiomyocytes. This might modify the expression of target genes by influencing the activity of transcription factors. To test this hypothesis, various investigations were performed. We found that overexpression of HSPA1A in HL-1 cells resulted in the differential expression of various genes. We used KEGG pathway and GO analysis to analyze these DEGs. We discovered that the upregulated genes were enriched in the coupled receptor of the G protein signaling, signal transduction, transportation, and other biological processes. The upregulation of many ncRNAs was also observed. After listing these ncRNAs according to expression, the top ncRNAs (Rn7SK, Rmrp) have been reported to be related to cardiac hypertrophy (49). The RNA-seq assessment and qRT-PCR verification of these ncRNAs (Rn7SK, Rmrp) are depicted in Figure 3. Upregulation of Rn7SK and Rmrp in pulmonary hypertension has also been observed. Moreover, we discovered that the downregulated genes were enriched in inflammatory and immune responses. Inflammation and immunity are associated with the pathogenesis of cardiac hypertrophy, specifically, the genes CXCL1, CCL2, CCL7, CXCL5, Fas, and C3 identified in the present study. After listing the downregulated DEGs from high to low, the top-ranking genes (CXCL1, CCL2, CCL7, CXCL5, Fas, and C3) were all associated with cardiac hypertrophy. The RNA-seq assessment and qRT-PCR verification of these genes are depicted in Figure 4.
Following the overexpression of HSPA1A, we identified 366 variable splicing events. The most relevant functions associated with these events were transcription and transcriptional regulation. Since the literature reports that HSPA1B can bind RNA and HSP72 is able to enhance the variable splicing of genes (33), we predict that HSPA1A may also regulate gene expression by promoting alternative transcription. To test this hypothesis, we carried out a series of experiments. It has been reported that HSPA1A regulates the alternative splicing of multiple genes (50). In our study, GO analysis showed that genes with substantial modifications in alternative splicing after HSPA1A overexpression were enriched in transcription, regulatory transcription, RNA splicing, DNA repair, mRNA processing, chromatin modification, regulatory translation, transport of calcium ion transmembrane, negative regulation of transcription of the promoter related to the RNA polymerase II, and DNA damage stimulation response. Several genes, including ASXL2 and RUNX1 enriched in these transcriptional regulatory pathways, have been reported to be associated with myocardial hypertrophy. The results were confirmed by RT-PCR verification, RNA-seq quantification, and ASE assessments, as shown in Figure 5. We also observed the upregulation of Runx1 and downregulation of Asxl2 in pathological cardiac hypertrophy which was as expected. All the materials and methods we used in our research provide a deep and thorough comprehension of the pathogenesis of pathological cardiac hypertrophy at both qualitative and quantitative levels.
A total of four groups of mouse ventricular tissue samples were established, and high-throughput sequencing was performed. The results suggested a hypothesis that HSPA1A may induce the development of cardiac hypertrophy through promoting or inhibiting the expression of ncRNAs related to cardiac hypertrophy and the immune-inflammatory response-related pathway genes. Non-coding RNAs do not translate into protein after the genome is transcribed. They include small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), microRNA (miRNA), piwi-interacting RNA (piRNA), small interfering RNA (siRNA), and long non-coding RNA (lncRNA).
In the process of AS, the exons of the RNA produced by transcription of the main gene or mRNA precursor are reconnected by RNA splicing (selecting different splicing sites) in a variety of ways. The resulting different mRNAs may be translated into different proteins. Therefore, a gene may encode multiple proteins. It has been shown that AS regulates cellular apoptosis as well as the subcellular localization of apoptosis-related gene products, the function and activity of apoptosis-related genes. Variable splicing is an important means of regulating gene expression and producing proteome diversity and is the main reason for the difference in the number of eukaryotic genes and proteins.
Taken together, this research indicates that the heat shock protein HSPA1A regulates the expression of immune and inflammatory genes or ncRNAs, possibly by modulating alternative splicing, which potentially participates in the occurrence and development of cardiac hypertrophy. In addition, the RNA-binding protein Zfp36 was significantly differentially expressed in both TAC + dantrolene vs. sham + dantrolene and the TAC + DMSO vs. sham + DMSO groups. These results suggest that Zfp36 may be also related to cardiac hypertrophy. The expression of mki67 and gm5619 was significantly different between the TAC + dantrolene group and the TAC + DMSO group. These results suggest that dantrolene may delay cardiac hypertrophy and ventricular remodeling and improve heart failure by regulating the expression of the RNA-binding protein genes MKI67 and GM5619. These results further expand the understanding of its strategy as a clinical therapeutic objective and suggest the possibility of developing treatments for cardiac hypertrophy and related maladies. Over the years, both the incidence and mortality of cardiac hypertrophy have remained high. The prevention and treatment of heart failure is a major challenge faced by cardiologists in the twenty-first century. Many studies have shown that ventricular remodeling is the basic mechanism of heart failure, and cardiac hypertrophy in ventricular remodeling is an independent risk factor of cardiovascular disease. Therefore, reversing cardiac hypertrophy would have profound significance in preventing the development of heart failure and reducing mortality. This study suggests a potentially effective prospect for disease treatment. At present, there is still a lack of effective clinical treatment of cardiac hypertrophy despite numerous cardiology treatments and drugs. A major contributing factor to this situation is a lack of full understanding of the pathophysiological process of cardiac hypertrophy and the molecular mechanisms involved in its development. The targeted therapy for myocardial hypertrophy and heart failure is not accurate. Now the clinical treatment is mostly aimed at improving symptoms, which cannot be corrected for the fundamental changes of the disease.
In this study, the RNA-binding protein HSPA1A was found to regulate the expression of genes related to cardiac hypertrophy, related ncRNAs, and immune and inflammatory response-related genes, as well as regulating the alternative splicing of genes related to transcriptional regulation in the development of cardiac hypertrophy. Further research is required into the transcriptional and posttranscriptional regulation of genes to fully understand the complex molecular mechanisms involved in cardiac hypertrophy to prevent and treat both the condition and related diseases.
Conclusions
The expression of HSPA1A was significantly upregulated in TAC mice, indicating an association of the gene with cardiac hypertrophy. It may be that Zfp36 is also related to cardiac hypertrophy. Dantrolene may delay cardiac hypertrophy and ventricular remodeling and improve heart failure by regulating the expression of the RNA-binding protein genes MKI67 and GM5619. The HSPA1A gene regulated the transcriptome of HL-1 cells, of which 69 genes were found to be significantly upregulated and 52 genes downregulated. In addition, HSPA1A selectively regulated the expression of ncRNAs (RN7SK and RMRP) related to cardiac hypertrophy. It negatively regulated the expression of inflammation and immune response-related genes CXCL1, CCL2, CCL7, CXCL5, FAS, C3, and LCN2. It also appears that HSPA1A plays a role in cardiac hypertrophy by negative regulation of AS of ASXL2 and positively regulating RUNX1 AS.
Acknowledgments
We thank Huan Sun, Xuelian Wang, and Fengyun Yu for their expert technical assistance.
Funding: This study was supported by grants provided by the National Natural Science Foundation of China (81970209).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-1222
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-1222
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-1222). Both authors report that this study was supported by grants provided by the National Natural Science Foundation of China (81970209). Both authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures were approved by the Institutional Animal Care and Use Committee of Jilin University and conducted in accordance with the Guideline for Ethical Review of Animal Welfare and Guide for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol 2016;97:245-62. [Crossref] [PubMed]
- Weeks KL, McMullen JR. The athlete's heart vs. the failing heart: can signaling explain the two distinct outcomes? Physiology (Bethesda) 2011;26:97-105. [Crossref] [PubMed]
- Nakamura M, Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol 2018;15:387-407. [Crossref] [PubMed]
- Burchfield JS, Xie M, Hill JA. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation 2013;128:388-400. [Crossref] [PubMed]
- Frieler RA, Mortensen RM. Immune cell and other noncardiomyocyte regulation of cardiac hypertrophy and remodeling. Circulation 2015;131:1019-30. [Crossref] [PubMed]
- Chen Y, Zhang Z, Zhu D, et al. Knockdown of KCNQ1OT1 attenuates cardiac hypertrophy through modulation of the miR-2054/AKT3 axis. J Thorac Dis 2020;12:4771-80. [Crossref] [PubMed]
- Xu L, Brink M. mTOR, cardiomyocytes and inflammation in cardiac hypertrophy. Biochim Biophys Acta 2016;1863:1894-903. [Crossref] [PubMed]
- Shen Y, Zhang X, Li C, et al. Pressure overload promotes cystatin C secretion of cardiomyocytes to regulate the MAPK signaling pathway and mediate cardiac hypertrophy. Ann Transl Med 2020;8:1514. [Crossref] [PubMed]
- Nomura S, Satoh M, Fujita T, et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun 2018;9:4435. [Crossref] [PubMed]
- Genest O, Wickner S, Doyle SM. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J Biol Chem 2019;294:2109-20. [Crossref] [PubMed]
- de Jong PR, Schadenberg AW, Jansen NJ, et al. Hsp70 and cardiac surgery: molecular chaperone and inflammatory regulator with compartmentalized effects. Cell Stress Chaperones 2009;14:117-31. [Crossref] [PubMed]
- Rosenzweig R, Nillegoda NB, Mayer MP, et al. The Hsp70 chaperone network. Nat Rev Mol Cell Biol 2019;20:665-80. [Crossref] [PubMed]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 2005;62:670-84. [Crossref] [PubMed]
- Żwirowski S, Kłosowska A, Obuchowski I, et al. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. EMBO J 2017;36:783-96. [Crossref] [PubMed]
- Morán Luengo T, Mayer MP, Rüdiger SGD. The Hsp70-Hsp90 Chaperone Cascade in Protein Folding. Trends Cell Biol 2019;29:164-77. [Crossref] [PubMed]
- Fernández-Fernández MR, Gragera M, Ochoa-Ibarrola L, et al. Hsp70 - a master regulator in protein degradation. FEBS Lett 2017;591:2648-60. [Crossref] [PubMed]
- Knapinska AM, Gratacós FM, Krause CD, et al. Chaperone Hsp27 modulates AUF1 proteolysis and AU-rich element-mediated mRNA degradation. Mol Cell Biol 2011;31:1419-31. [Crossref] [PubMed]
- Sinsimer KS, Gratacós FM, Knapinska AM, et al. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol Cell Biol 2008;28:5223-37. [Crossref] [PubMed]
- Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA 2010;1:457-73. [Crossref] [PubMed]
- Hulina A, Grdić Rajković M, Jakšić Despot D, et al. Extracellular Hsp70 induces inflammation and modulates LPS/LTA-stimulated inflammatory response in THP-1 cells. Cell Stress Chaperones 2018;23:373-84. [Crossref] [PubMed]
- Martine P, Chevriaux A, Derangère V, et al. HSP70 is a negative regulator of NLRP3 inflammasome activation. Cell Death Dis 2019;10:256. [Crossref] [PubMed]
- Kumada K, Fuse N, Tamura T, et al. HSP70/DNAJA3 chaperone/cochaperone regulates NF-κB activity in immune responses. Biochem Biophys Res Commun 2019;513:947-51. [Crossref] [PubMed]
- Willis MS, Min JN, Wang S, et al. Carboxyl terminus of Hsp70-interacting protein (CHIP) is required to modulate cardiac hypertrophy and attenuate autophagy during exercise. Cell Biochem Funct 2013;31:724-35. [Crossref] [PubMed]
- Di Naso FC, Porto RR, Fillmann HS, et al. Obesity depresses the anti-inflammatory HSP70 pathway, contributing to NAFLD progression. Obesity (Silver Spring) 2015;23:120-9. [Crossref] [PubMed]
- Chung J, Nguyen AK, Henstridge DC, et al. HSP72 protects against obesity-induced insulin resistance. Proc Natl Acad Sci U S A 2008;105:1739-44. [Crossref] [PubMed]
- Gehrig SM, van der Poel C, Sayer TA, et al. Hsp72 preserves muscle function and slows progression of severe muscular dystrophy. Nature 2012;484:394-8. [Crossref] [PubMed]
- Wei YQ, Zhao X, Kariya Y, et al. Inhibition of proliferation and induction of apoptosis by abrogation of heat-shock protein (HSP) 70 expression in tumor cells. Cancer Immunol Immunother 1995;40:73-8. [Crossref] [PubMed]
- Kumar S, Stokes J 3rd, Singh UP, et al. Targeting Hsp70: A possible therapy for cancer. Cancer Lett 2016;374:156-66. [Crossref] [PubMed]
- Sherman MY, Gabai VL. Hsp70 in cancer: back to the future. Oncogene 2015;34:4153-61. [Crossref] [PubMed]
- Meng L, Hunt C, Yaglom JA, et al. Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene 2011;30:2836-45. [Crossref] [PubMed]
- Kim YK, Suarez J, Hu Y, et al. Deletion of the inducible 70-kDa heat shock protein genes in mice impairs cardiac contractile function and calcium handling associated with hypertrophy. Circulation 2006;113:2589-97. [Crossref] [PubMed]
- Kee HJ, Eom GH, Joung H, et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ Res 2008;103:1259-69. [Crossref] [PubMed]
- Hahn A, Bublak D, Schleiff E, et al. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011;23:741-55. [Crossref] [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [Crossref] [PubMed]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511-5. [Crossref] [PubMed]
- Jin L, Li G, Yu D, et al. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus Verticillium dahliae. BMC Genomics 2017;18:130. [Crossref] [PubMed]
- Wolff A, Bayerlová M, Gaedcke J, et al. A comparative study of RNA-Seq and microarray data analysis on the two examples of rectal-cancer patients and Burkitt Lymphoma cells. PLoS One 2018;13:e0197162 [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621-8. [Crossref] [PubMed]
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316-W322. [Crossref] [PubMed]
- Kitamura Y, Mise N, Mori Y, et al. Proteomic identification of the proteins related to cigarette smoke-induced cardiac hypertrophy in spontaneously hypertensive rats. Sci Rep 2020;10:18825. [Crossref] [PubMed]
- Marques FZ, Prestes PR, Byars SG, et al. Experimental and Human Evidence for Lipocalin-2 (Neutrophil Gelatinase-Associated Lipocalin NGAL) in the Development of Cardiac Hypertrophy and heart failure. J Am Heart Assoc 2017;6:005971 [Crossref] [PubMed]
- Flo TH, Smith KD, Sato S, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004;432:917-21. [Crossref] [PubMed]
- Lai HL, Grachoff M, McGinley AL, et al. Maintenance of adult cardiac function requires the chromatin factor Asxl2. J Mol Cell Cardiol 2012;53:734-41. [Crossref] [PubMed]
- Baskind HA, Na L, Ma Q, et al. Functional conservation of Asxl2, a murine homolog for the Drosophila enhancer of trithorax and polycomb group gene Asx. PLoS One 2009;4:e4750 [Crossref] [PubMed]
- Brunner R, Lai HL, Deliu Z, et al. Asxl2-/- Mice Exhibit De Novo Cardiomyocyte Production during Adulthood. J Dev Biol 2016;4:32. [Crossref] [PubMed]
- Gattenlöhner S, Waller C, Ertl G, et al. NCAM(CD56) and RUNX1(AML1) are up-regulated in human ischemic cardiomyopathy and a rat model of chronic cardiac ischemia. Am J Pathol 2003;163:1081-90. [Crossref] [PubMed]
- McCarroll CS, He W, Foote K, et al. Runx1 Deficiency Protects Against Adverse Cardiac Remodeling After Myocardial Infarction. Circulation 2018;137:57-70. [Crossref] [PubMed]
- Greco S, Zaccagnini G, Perfetti A, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med 2016;14:183. [Crossref] [PubMed]
- Qin K, Jian D, Xue Y, et al. DDX41 regulates the expression and alternative splicing of genes involved in tumorigenesis and immune response. Oncol Rep 2021;45:1213-25. [Crossref] [PubMed]
(English Language Editor: J. Jones)


