
Resveratrol inhibits cell apoptosis by suppressing long noncoding RNA (lncRNA) XLOC_014869 during lipopolysaccharide-induced acute lung injury in rats
Introduction
Acute inflammation of the alveoli and lung parenchyma is a part of pathogenic mechanisms in the common clinical syndrome acute lung injury (ALI) (1). ALI can be induced by many factors, such as sepsis, trauma, endotoxemia, and inhalation of harmful gases (2). An uncontrolled inflammatory response occurs during ALI pathogenesis. Pro-inflammatory cells and the cytokines they release can cause lung dysfunction (3). Although there are many treatment, the mortality rate of ALI patients is still high (4). It is therefore, necessary to clarify the mechanisms of ALI and find new prevention and treatment to reduce morbidity and mortality.
Resveratrol (Res) is a polyphenolic plant antitoxin with antioxidant, anticancer, and anti-inflammatory activities (5,6). A review has reported that Res has protective and therapeutic effects in the setting of ALI. Res can protect against ALI induced by sepsis by activating the vascular endothelial growth factor-β signaling pathway (7). Oral administration of Res-loaded lipid-core nanocapsules before intranasal infusion of lipopolysaccharide (LPS) can improve mouse lung function and prevent the accumulation of total leukocytes and neutrophils in bronchoalveolar fluid (BALF) and lung tissues (8). Res also has a protective effect on LPS-induced lung injury in mice by inhibiting NLP family pyrin domain-containing 3 inflammasome formation (9). From this point of view, results on the mechanism by which Res protects against ALI are not consistent.
Long noncoding RNAs (lncRNAs) are the largest type of noncoding RNA produced by the genome. At present, there are 16,193 annotated lncRNAs in the human genome, and <3% of these have known corresponding functions (10). It seems that lncRNAs can regulate gene expression at transcriptional and post-transcriptional levels through a variety of mechanisms (11). Whether lncRNA is involved in Res-mediated protection against ALI is unknown, underscoring the need for further study into the role of lncRNAs in protection from lung injury.
This study aimed to explore the protective mechanism of Res on ALI and the role of lncRNAs in this protective process by using rat model of ALI and LPS-treated alveolar type II cells. Our results build on those published by other researchers describing the protective effect of Res against ALI and provide a deeper understanding of the underlying mechanism.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-1113).
Methods
Rat model of LPS-induced ALI
Twenty male Sprague-Dawley rats (7–8 weeks old) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). All the rats were kept in pathogen-free housing under a 12-h light/dark cycle with free access to food and sterile water during the research procedures. The rats were anesthetized with 3% sodium pentobarbital (150 mg/kg), and randomly divided into four groups (five rats per group): control (K group): intraperitoneal injection of physiological saline; LPS (L group): intraperitoneal injection of 200 µL of LPS (5 mg/kg; Sigma, St. Louis, MO, USA) dissolved in physiological saline; LPS + dexamethasone (LPS + Dexa group, positive control): intragastric administration of Dexa (1.5 mg/kg) (Sigma) 1 h before intraperitoneal injection of LPS (5 mg/kg); LPS + Res (R group in sequencing data): intragastric administration of Res (0.5 mg/kg) 1 h before intraperitoneal injection of LPS (5 mg/kg). At 6 h after LPS treatment, the rats were euthanized by sodium pentobarbital injection (200 mg/kg). All experimental procedures were approved by the ethics committee of Shanghai Pulmonary Hospital (approval No. K21-025), in compliance with National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
BALF collection
The thorax was opened, the left lung tissues and trachea were taken out, and 2 mL saline solution was introduced into the trachea for three washes, a total of 6 mL washing liquid was collected. The sample was centrifuged at 2,500 ×g for 10 min, and the supernatant was collected and stored at −80 °C, total protein in BALF was detected using Bradford assays.
Lung edema measurement
Lung edema was determined by calculating the lung wet/dry weight ratio. Briefly, the middle lobes of the right lungs were removed and weighed to obtain the wet weight. The tissues were then dried at 95 °C for 24 h and reweighed to obtain the dry weight. The wet-to-dry ratio was calculated by dividing the wet weight by the dry weight.
Pathological studies
Lung tissues were fixed with 4% paraformaldehyde (Servicebio, Wuhan, China), dehydrated with graded ethanol until transparent, waxed, and embedded in sequence, and then cut into 5-µm sections using a microtome. The samples were stained with hematoxylin and eosin (H&E) (LTD) to study histopathological changes. Lung injury was evaluated according to a semiquantitative scoring system assessing alveolar congestion, hemorrhage, infiltration or aggregation of neutrophils in airspaces or vessel walls, and thickness of alveolar wall/hyaline membrane formation, and inflammatory cell infiltration (12). The results were graded from 0 to 4 for each variable, and the total score representing lung injury ranged from 0 to 16 (13).
Nitric oxide (NO) determination
Serum was separated by centrifuging at 1,500 rpm for 10 min at 4 °C after blood samples were allowed to stand at room temperature for 2 h. The concentrations of NO in the serum and BALF were measured using Griess reagent in NO assay kits (S0021S, Beyotime, Shanghai, China) according to the manufacturer’s instructions.
Enzyme-linked immunosorbent assay (ELISA)
The levels of tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-1β, IL-8, and interferon (IFN)-γ in BALF, serum, and lung tissue were measured using specific ELISA kits according to the manufacturer’s instructions (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China).
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay
TUNEL assay kits were purchased from Servicebio and used according to the manufacturer’s instructions. The main steps included antigen retrieval on dewaxed tissue sections using proteinase K working solution at 37 °C for 25 min, permeabilization using permeabilize working solution at room temperature for 20 min, incubation in a reaction solution composed of TDT enzyme, dUTP and buffer at 37 °C for 2 h; and nuclei counterstaining with 4',6-diamidino-2-phenylindole. The sections were observed under a fluorescence microscope (NIKON ECLIPSE C1, Tokyo, Japan) and images were collected using the imaging system (NIKON DS-U3).
RNA extraction and lncRNA sequencing
Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) following the manufacturer’s protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with RNA integrity number ≥7 were subjected to the subsequent analysis. The libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold according to the manufacturer’s instructions. Then these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500, Illumina, San Diego, CA, USA) and 150-/125-bp paired-end reads were generated.
Bioinformatics analysis
To obtain high-quality reads that could be used for later analysis, raw reads were quality filtered. Trimmomatic (14) software was first used for adapter removing, and then low-quality bases and N-bases or low-quality reads were filtered out to yield high-quality clean reads. Using hisat2 (15) to align clean reads to the reference genome, the samples were assessed by genomic and gene alignment. The results of alignment with the reference genome were stored in a binary file, called a bam file. Using Stringtie (16) software to assemble the reads, the new transcript was spliced, then the candidate lncRNA transcripts were selected by comparing the gene annotation information of the reference sequence produced by Cuffcompare (17) software. Finally, transcripts with coding potential were screened out by CPC (18), CNCI (19), Pfam (20), and PLEK (21) to obtain lncRNA predicted sequences. The sequencing reads of each sample were aligned with mRNA transcript sequences, known lncRNA sequences, and lncRNA prediction sequences by bowtie2. The fragments per kilobase of exon per million mapped fragment values and count values (the number of reads for each gene in each sample) were obtained by gene quantitative analysis using eXpress. The estimateSizeFactors function of the DESeq (22) R package was used to normalize the counts, and the nbinomTest function was employed to calculate P values and foldchange (FC) values for the difference comparison. We selected differential transcripts with P values ≤0.05 and |FC| >2, and analyzed differentially expressed mRNAs following Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment using hypergeometric distribution tests.
Cell culture and treatment
Rat lung alveolar type II cells were purchased from Shanghai FuHeng BioLogy Co., Ltd. (Shanghai, China), and cultured in M199 complete medium containing 10% fetal bovine serum and epithelial cell growth factor (Thermo Fisher Scientific, Waltham, MA, USA). The cells were divided into four groups to assess the effect of Res on LPS-treated alveolar cells: control, LPS (10 µg/mL), Res (30 µM) + LPS (10 µg/mL), and Dexa (0.1 µM) + LPS (10 µg/mL). For the latter two groups, cells were treated with Res or Dexa for 24 h and then with LPS for 12 h. Cells were seeded in a 24-well plate (1×105 cells/well) to detect the efficiency of short interfering RNAs (siRNAs) against lncRNA XLOC_014869. The sequences of si-XLOC_014869 (si-869) and si-NC were as follows: si-869-1 sense: 5'-GCC CUG UGC AAA GUA UAU ATT-3', antisense: 5'-UAU AUA CUU UGC ACA GGG CTT-3'; si-869-2 sense: 5'-GCC CAG GGA AAU GUU UCA ATT-3', antisense: 5'-UUG AAA CAU UUC CCU GGG CTT-3'; si-869-3 sense: 5'-GCU GCC UUU AGC CAA UCA ATT-3', antisense: 5'-UUG AUU GGC UAA AGG CAG CTT-3'; si-NC sense: 5'-UUC UCC GAA CGU GUC ACG UTT-3', antisense: 5'-ACG UGA CAC GUU CGG AGA ATT-3'. For RNA interference assay, the cells were divided into five groups: control, LPS, Res + LPS, si-NC + LPS, and si-869 + LPS.
Quantitative real-time polymerase chain reaction (PCR)
Extraction of total RNA from lung tissue and cells was performed with TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and further reverse-transcribed using 5× Prime Script RT Master Mix kit (Takara, Kusatsu, Japan). SYBR qPCR Master Mix (Takara) was used to carry out quantitative PCR according to the manufacturer’s instructions. Target gene expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels in respective samples as an internal control and calculated using the 2−ΔΔCt method. The primer sequences are shown in Table 1.
Table 1
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| Fos | TTTCAACGCGGACTACGAGG | TCGGCTGGGGAATGGTAGTA |
| Fosb | GCCGAGTCTCAGTACCTGTC | GGTCCTGGCTGGTTGTGATT |
| Jun | GCATAGCCAGAATACGCTGC | GTTGCTGAGGTTGGCGTAGA |
| Faslg | TGGTGGCTCTGGTTGGAATG | CTCACGGAGTTCTGCCAGTT |
| XLOC_001387 | TACAGCGGAGGTCTGAAGG | GTGGGATGAGATGCGAGT |
| XLOC_014869 | TTGTCAAGATGGGTGGGT | CAGGGCAGGCAGGTAGAA |
| GAPDH | GAAGGTCGGTGTGAACGGAT | ACCAGCTTCCCATTCTCAGC |
PCR, polymerase chain reaction; lncRNA, long noncoding RNA.
Western blotting
Tissues and cells were lysed with radioimmunoprecipitation assay buffer (Beyotime) containing complete protease inhibitor. The total protein levels were quantified by Bradford assays. Equal amounts of protein (30 µg) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk for 2 h and probed with the primary antibodies diluted in blocking solution overnight at 4 °C. Then the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. Immunoreactive bands were observed by using a gel imaging system with an enhanced chemiluminescence system (Thermo Fisher Scientific). Primary antibodies against c-Jun (AF6090), phospho-c-Jun (AF3095), Fas-L (AF5333), c-Fos (AF0132), and GAPDH (AB0037) were purchased from Affinity Biosciences (Cincinnati, OH, USA) and Abways (Shanghai, China), respectively.
Flow cytometry
The cells were collected and washed with precooled phosphate-buffered saline twice and centrifuged at 1,000 ×g each at 4 °C for 5 min after each wash. The cells were then resuspended in 195 µL 1× binding buffer and incubated with 5 µL Annexin-V-FITC and 10 µL PI contamination solution (C1062M, Beyotime) in the dark for 15 min for double staining. The samples were subjected to flow cytometry (FACStar, BD Biosciences, Foster City, CA, USA). The percentage of apoptotic cells was analyzed using CellQuest software (BD Biosciences).
Statistical analysis
All data were analyzed using GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). Repeated-measure data are presented as means ± standard deviations (SDs) and were statistically analyzed using repeated-measures analysis of variance (ANOVA). P<0.05 was considered statistically significant.
Results
Res alleviated histological injury in LPS-induced ALI
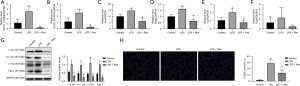
Tissues from all 20 rats were sampled for pathological and injury index detection. H&E staining showed that LPS induced injury in rat lungs, but the degree of injury was significantly less in the LPS + Res group compared to the LPS group, and similar to that in LPS + Dexa group (Figure 1A). Evaluation of lung injury according to the semiquantitative scoring system also showed that lung injury was more severe in the LPS group (10.00 vs. 5.60 in the LPS + Res group, P<0.01) (Figure 1B). In BALF, LPS induced high levels of total protein compared with the control group, while Res and Dexa suppressed the increase of total protein in the LPS + Res group and LPS + Dexa group respectively (Figure 1C). Furthermore, LPS induced high serum levels of NO compared with the control group, while Res suppressed the increase of NO in the LPS + Res group (Figure 1D). There were no significant differences in BALF NO concentrations or the wet/dry lung ratio (Figure 1E,1F).
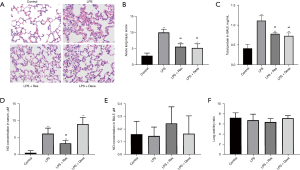
Res decreased the inflammatory cytokines in BALF, serum, and lung tissues
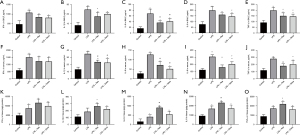
The levels of TNF-α, IL-6, IL-1β, IL-8, and IFN-γ in BALF, serum, and lung tissue were measured using specific ELISA kits. All five cytokines were significantly increased in the BALF, serum, and lung tissues of rats treated with LPS. In BALF, Res pretreatment attenuated the increases of IL-1β, IL-6, and TNF-α, while Dexa pretreatment dampened the increases of IL-6, IL-8 and TNF-α. Res or Dexa pretreatment inhibited the increases of IL-1β, IL-6, IL-8, and TNF-α in serum. Res pretreatment enhanced IL-1β, IL-6, IL-8, and TNF-α in lung tissue (Figure 2).
LPS and Res altered lncRNA and mRNA profile in lung
Through lncRNA sequencing, we detected an average of 17,292 mRNAs and 6,812 lncRNAs in each sample. We compared in the three groups using P<0.05 and |FC| >2 as the screening criteria to screen differentially expressed lncRNAs and mRNAs (https://cdn.amegroups.cn/static/public/jtd-21-1113-1.xlsx). Compared with the control group, the number of differentially expressed mRNAs in the LPS and LPS + Res groups were 4,112 and 4,242, respectively; while there were 101 differentially expressed mRNAs in Res + LPS group compared with LPS group (Figure 3A-3C). There were 5,032 known lncRNAs, and 1,780 novel lncRNAs (Table 2). Through prediction analysis of the coding ability of lncRNAs using CPC, CNCI, PFAM and PLAEK software, we obtained 6,773, 6,443, 6,716, and 5,633 non-coding potential lncRNAs, respectively, and 4,252 lncRNAs showed non-coding potential in the prediction results of the four software programs (Figure 3D). Compared with the control group, the numbers of upregulated lncRNAs in the LPS and Res + LPS groups were 570 and 574, and the numbers of down-regulated lncRNAs were 640 and 627, respectively (Figure 3E,3F); compared with the LPS group, the numbers of up and downregulated lncRNAs in the Res + LPS group were 97 and 84, respectively (Figure 3G).
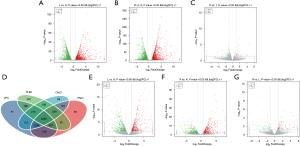
Table 2
| Sample | mRNA | lncRNA | ||
|---|---|---|---|---|
| Known | Predict | All | ||
| K_2 | 17,468 | 5,349 | 1,777 | 7,126 |
| K_3 | 17,563 | 5,501 | 1,777 | 7,278 |
| K_5 | 17,362 | 5,453 | 1,766 | 7,219 |
| L_1 | 17,219 | 4,732 | 1,778 | 6,510 |
| L_3 | 17,172 | 4,985 | 1,786 | 6,771 |
| L_5 | 17,144 | 5,044 | 1,784 | 6,828 |
| R_2 | 17,255 | 4,735 | 1,776 | 6,511 |
| R_3 | 17,257 | 4,768 | 1,796 | 6,564 |
| R_5 | 17,186 | 4,721 | 1,784 | 6,505 |
lncRNAs, long noncoding RNAs.
Differentially expressed mRNAs and lncRNAs involved in immunity, infection, and signal transduction
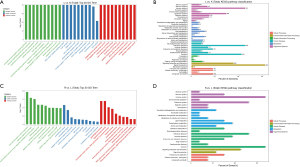
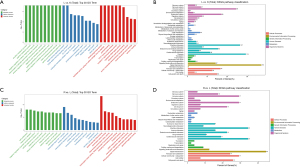
After identifying the differential mRNAs, we performed GO enrichment analysis to compare the LPS and control groups, and LPS + Res and LPS groups. The KEGG database was used to perform pathway analysis. Compared with the control group, differentially expressed mRNAs in the LPS group were mainly located in the cell membrane and outside the cell and, involved in protein binding and receptor binding functions, participating in the immune process, stress response, and cytokines (Figure 4A). The KEGG signaling pathways were mainly involved in immunity, infection, and signal transduction (Figure 4B). Compared with the LPS group, proteins encoded by differentially expressed mRNAs in the LPS + Res group were mainly located in the extracellular, cell membrane, and cell connections, exerting nucleic acid binding and enzyme activation, participating in immunity, responds to stimulation, and signal transduction (Figure 4C). The KEGG signaling pathways were mainly involved in immunity, infection, signaling molecules and interactions (Figure 4D). Subsequent GO enrichment analysis and pathway analysis were performed on partner genes of differentially expressed lncRNAs. Compared with the control group, the partner genes of lncRNAs in the LPS group were mainly located in the complex, played a binding and activation function, and participated in biological regulation and cellular processes (Figure 5A). The KEGG signaling pathways were mainly involved in immunity, infection, and signal transduction (Figure 5B). Compared with the LPS group, partner genes in the LPS + Res group were mainly located in extracellular, cell membrane, and cell connection, played an activation function; and participated in immune and regulatory processes (Figure 5C). The KEGG analysis of adjacent genes of lncRNAs between the LPS and LPS + Res groups revealed three main signaling pathways: immunity, infection, and signal transduction (Figure 5D).
Res inhibited the increase of lncRNAs TCONS_00033096 (XLOC_014869) and TCONS_00003069 (XLOC_001387) in LPS-treated rat lungs
Using P<0.05 and |FC| >2 as the screening criteria, we further compared differentially expressed mRNAs and lncRNAs between the LPS and control groups and LPS + Res and LPS groups. There were 26 and 23 differentially expressed mRNAs and lncRNAs, respectively, with higher levels in the LPS group that were decreased in LPS + Res group, respectively; there were 17 and 27 differentially expressed mRNAs and lncRNAs, respectively, with lower levels in the LPS group that increased in the LPS + Res group (Figure 6A,6B). Comparing the mRNAs with the partner genes of the lncRNAs, we found that the partner genes Fos and Fosb of lncRNAs TCONS_00033096 (XLOC_014869) and TCONS_00003069 (XLOC_001387) increased in the LPS group but decreased in the LPS + Res group (Table 3). KEGG analysis showed that Fos and Fosb participate as transcription factors in a variety of signaling pathways, including apoptosis, Toll-like receptor signaling, and TNF signaling. We therefore, analyzed the expression of the downstream proteins regulated by Fos and Fosb in the LPS and LPS + Res groups. The results showed that the mRNA levels of Jun and Faslg with pro-apoptotic function increased in the LPS group but decreased in the LPS + Res group (Table 4).
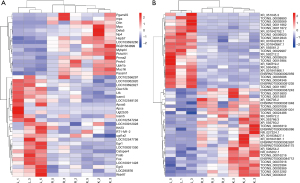
Table 3
| lncRNA_id | lncRNA_symbol | partnerRNA_transcript | partnerRNA_gene |
|---|---|---|---|
| ENSRNOT00000081048 | AABR07035005.1 | XM_008769022.2 | LOC102557071 |
| ENSRNOT00000082897 | AABR07045680.1 | NM_001100804.1 | Cd99 |
| ENSRNOT00000084175 | AABR07054126.1 | XM_006235140.3 | Foxa2 |
| ENSRNOT00000084712 | AABR07001382.1 | XM_017590500.1 | Syne1 |
| ENSRNOT00000085086 | AABR07065895.1 | XM_006240685.3 | Zfp386 |
| ENSRNOT00000087243 | AABR07011013.1 | XM_008761072.2 | LOC365085 |
| ENSRNOT00000091235 | AABR07065118.1 | NM_001037783.2 | Lrrc74a |
| ENSRNOT00000091301 | AABR07065448.1 | XM_003750226.4 | LOC100909476 |
| ENSRNOT00000092959 | AABR07035955.1 | XM_008769176.1 | LOC100912379 |
| NR_027324.1 | H19 | XM_008760183.2 | LOC102547221 |
| NR_045202.1 | Tmem80 | NM_031801.2 | Deaf1 |
| TCONS_00001008 | XLOC_000482 | NM_001106267.1 | Tm2d3 |
| TCONS_00003069 | XLOC_001387 | NM_001256509.1 | Fosb |
| TCONS_00005369 | XLOC_002444 | XM_340803.7 | Sap30l |
| TCONS_00008890 | XLOC_004012 | NM_001105881.2 | Naa50 |
| TCONS_00009523 | XLOC_004304 | XM_017598594.1 | Srrt |
| TCONS_00009526 | XLOC_004304 | XM_017598594.1 | Srrt |
| TCONS_00011862 | XLOC_005379 | NM_001109084.1 | Fam71a |
| TCONS_00011867 | XLOC_005380 | XM_017599013.1 | LOC100911428 |
| TCONS_00011872 | XLOC_005380 | NM_012912.2 | Atf3 |
| TCONS_00012282 | XLOC_005537 | XM_017599544.1 | LOC108352829 |
| TCONS_00012843 | XLOC_005775 | XM_006250632.3 | Aff1 |
| TCONS_00015800 | XLOC_007130 | XM_006253206.3 | Msr1 |
| TCONS_00015801 | XLOC_007130 | NM_001191939.1 | Msr1 |
| TCONS_00015984 | XLOC_007207 | NM_001135009.1 | Col4a1 |
| TCONS_00016216 | XLOC_007317 | XM_006253786.3 | Nedd9 |
| TCONS_00023394 | XLOC_010496 | XM_006256419.3 | Spock2 |
| TCONS_00024486 | |||
| TCONS_00029987 | |||
| TCONS_00030041 | XLOC_013484 | XM_006238139.3 | Tmem8b |
| TCONS_00033096 | XLOC_014869 | NM_022197.2 | Fos |
| TCONS_00033319 | XLOC_014979 | XM_017594191.1 | Gpr132 |
| TCONS_00034346 | XLOC_015440 | NM_001108726.1 | Itgb8 |
| TCONS_00037616 | XLOC_016901 | NM_022667.2 | Slco2a1 |
| XR_001838427.1 | LOC100911923 | NM_013093.1 | Nkx2-1 |
| XR_001840567.1 | LOC102552590 | XM_006248540.3 | Dgkg |
| XR_001841361.1 | LOC108348251 | NM_001012042.1 | Xkr6 |
| XR_001841866.1 | LOC102547930 | XM_017600453.1 | Edn1 |
| XR_001842358.1 | LOC103694359 | NM_001014271.1 | LOC367515 |
| XR_001842792.1 | LOC103690083 | XM_008773581.2 | LOC103690902 |
| XR_351045.3 | LOC102549247 | XM_008760362.2 | Ide |
| XR_353766.3 | LOC102553163 | NM_031046.3 | Itpr2 |
| XR_590319.2 | LOC102549305 | NM_012999.1 | Pcsk6 |
| XR_590795.2 | LOC102549789 | XM_008760627.2 | LOC684998 |
| XR_592212.2 | LOC102554570 | XM_006236915.3 | Adamts9 |
| XR_593561.2 | Pvt1 | NM_012603.2 | Myc |
| XR_593572.2 | Pvt1 | NM_012603.2 | Myc |
| XR_593780.2 | LOC102552294 | NM_001271048.2 | Cep295 |
| XR_595436.2 | LOC102550337 | NM_001271235.1 | Tnfrsf11a |
| XR_596254.2 | LOC102556284 | ||
lncRNAs, long noncoding RNAs.
Table 4
| Function | Gene | Protein | LPS vs. control | LPS + Res vs. LPS |
|---|---|---|---|---|
| Pro-apoptotic | Jun | c-Jun | Up | Down |
| Tp53 | p53 | Up | ||
| Fas | Fas | Up | ||
| Faslg | Fas-L | Up | Down | |
| Bcl2l11 | Bim | Up | ||
| Hrk | HRK | |||
| Cell survival | Ccnd1 | Cyclin D1 | Down | |
| Ccnd2 | Cyclin D2 | |||
| Ccnd3 | Cyclin D3 | Up | ||
| Tissue remodeling | Mmp1b | MMP1 | ||
| Mmp1 | MMP1 | |||
| Mmp3 | MMP3 | |||
| Mmp9 | MMP9 | Up | ||
| Mmp13 | MMP13 | Up | ||
| Chemokines | Cxcl1 | CXCL1 | Up | |
| Cxcl2 | CXCL2 | Up | ||
| Cxcl3 | CXCL3 | Up | ||
| Cxcl6 | CXCL5 | Up | ||
| Cxcl10 | CXCL10 | Up | ||
| Ccl2 | CCL2 | Up | Up | |
| Ccl12 | CCL2 | Up | Up | |
| Ccl7 | CCL7 | Up | Up | |
| Ccl20 | CCL20 | Up | ||
| Cytokines | Il6 | IL-6 | Up | |
| LOC103694380 | TNF | Up | ||
| Tnf | TNFα | Up | ||
| Ptgs2 | COX2 | Up | ||
| Csf3 | G-CSF | Up | ||
| Csf2 | GM-CSF | Up | ||
| Anti-microbial | Defb4 | Defensin beta 4 | ||
| Muc5ac | MUC5AC | Up | Up | |
| Muc5b | MUC5B | Up | ||
| S100a8 | S100A8 | Up | ||
| S100a9 | S100A9 | Up | ||
| Lcn2 | Lipocalin 2 | Up | ||
| Phagolysosome | Mpo | MPO | Down | Up |
| RT1-M1-2 | MHC1 | Up | Down | |
LPS, lipopolysaccharide; Res, resveratrol; IL, interleukin; TNF, tumor necrosis factor.
Res pretreatment inhibited apoptosis in LPS-treated rat lung cell
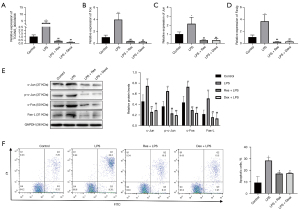
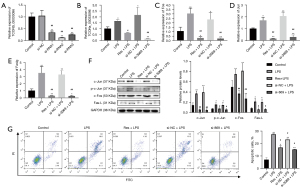
We detected the levels of XLOC_014869, XLOC_001387, Fos, Fosb, Faslg and Jun in lung tissues of the control, LPS, and LPS + Res groups by quantitative real-time PCR. The results verified higher levels of XLOC_014869, XLOC_001387, Fos, Faslg and Jun in the LPS group and lower levels in the LPS + Res group (Figure 7A). The protein levels of c-Fos, Fas-L, and c-Jun and the phosphorylation level of c-Jun also changed accordingly (Figure 7B). Compared with the control group, TUNEL results showed significantly increased apoptosis in the LPS group, which was effectively alleviated by Res pretreatment (Figure 7C).
Res pretreatment inhibited apoptosis of lung alveolar type II cells treated with LPS in vitro
Compared with the control group, the relative mRNA expression levels of genes in the LPS group were significantly increased. Compared with the LPS group, they were significantly decreased in the LPS + Res group (P<0.01) (Figure 8A). Western blotting showed that the relative protein levels of c-Jun, c-Fos and Fas-L and the phosphorylation level of c-Jun were significantly increased in the LPS group compared with the control group. Protein levels of c-Jun, c-Fos and Fas-L and the phosphorylation level of c-Jun were significantly decreased in the LPS + Res group compared to the LPS group (P<0.01) (Figure 8B). We observed increased apoptosis of lung alveolar type II cells in the LPS group compared to the control, LPS + Res, and LPS + Dexa groups (Figure 8C).
lncRNA XLOC_014869 knockdown reduced LPS-induced apoptosis of lung alveolar type II cells and c-Fos/c-Jun/Fas-L signaling
Quantitative real-time PCR showed that the three siRNAs all produced significant knockdown effects on lncRNA XLOC_014869, and the si-869-2 with ~80% knockdown was selected for subsequent experiments (Figure 9A). Compared with the alveolar type II cells treated with LPS alone, knockdown of lncRNA XLOC_014869 in LPS-treated cells yielded similar results to the cells treated by Res + LPS. The mRNA (Figure 9B) and protein expression levels (Figure 9C) of Fos, Fas-L and Jun were correspondingly downregulated. Flow cytometry confirmed that both Res and XLOC_014869 knockdown reduced apoptosis of LPS-induced alveolar type II cells (Figure 9D).
Discussion
We established an LPS-induced ALI rat model and performed in vitro experiments to explore the role of lncRNAs in the protective of Res. The findings indicated that pretreatment with Res reduced LPS-stimulated increases in XLOC_014869 and lung cell apoptosis. These effects may be related to XLOC_014869-mediated regulation of c-Fos/c-Jun/Fas-L axis, which mediates apoptosis in lung cells treated with LPS. Similar results were observed in rat lung alveolar type II cells.
During ALI pathogenesis, the release of inflammatory factors and mediators leads to the death of numerous alveolar epithelial cells (23). Controlling abnormal inflammation and apoptosis is conducive to improving prognosis (24). Pretreatment with Res reduced LPS-induced lung tissue damage and serum increases in NO, IL-1β, IL-6, IL-8, and TNF-α, and downregulated IL-1β, IL-6, and TNF-α in BALF of LPS-treated rats in this study. Given that IL-1β, IL-6, and TNF-α are pro-inflammatory cytokines (25), our results indicate a protective effect of Res against lung injury and inflammation (26). However, levels of these inflammatory factors in lung tissues from rats pretreated with Res were higher than compared to animals only given LPS. These inflammatory factors in lung tissues were not detected in similar studies (9,27), and it is unknown whether the reason is the dose and timing of Res treatment (28). This is an issue that deserves further study.
NO concentration in BALF and wet/dry lung ratio showed no significant changes, but BALF protein was abnormal between groups in our study. One group reported an increase in the concentration of NO in lung tissues and an increase in the wet/dry lung ratio after 4 h of LPS treatment, but the change of NO in BALF was not explained (29). However, another report showed that the wet/dry lung ratio or nitrites and total protein in BALF were increased significantly 6 h after LPS treatment (30). Our results were not completely consistent with those of existing studies, and further studies are also needed to confirm this.
In addition, differentially expressed lncRNAs and mRNAs in LPS-stimulated ALI lung tissue were mainly enriched in immune, stress and signal transduction-related pathways, this was similar to the results of another group (31). Meanwhile, we found that differentially expressed lncRNAs and mRNAs in Res-pretreated ALI lung tissue were also enriched in immune, stress and signal transduction related pathways. Most lncRNAs may activate or inhibit the expression of adjacent genes (32). In our study, Res reduced LPS-stimulated increases in lncRNAs XLOC_014869 and XLOC_001387 and the partner genes Fos and Fosb. Other differentially expressed lnRNAs were not associated with the differentially expressed mRNAs, and no similar studies have been reported. Therefore, the role of other differentially expressed lncRNAs and mRNAs in the protection of Res against ALI are still unknown.
Many studies have investigated the effects of lncRNA on apoptosis in lung diseases. The lncRNA small nucleolar RNA host gene 16 was highly expressed in patients with acute pneumonia and LPS-induced WI-38 cells, and was involved in LPS-induced apoptosis (33). Another group reported that lncRNA CASC2 was significantly reduced in LPS-induced A549 cells and ALI mouse models, while, CASC2 overexpression inhibited LPS-induced apoptosis of A549 cells (34). lncRNA MALAT1 is overexpressed in patients with acute respiratory distress syndrome may promote cell apoptosis by sponging miR-425 and participating in the pathophysiological process of lung damage (35). Studies on the protective mechanism of Res against ALI mainly focused on inflammation-related pathways (9). We examined lncRNA expression profiles in LPS-stimulated ALI and Res-pretreated ALI lung tissues to understand the role of lncRNA in Res-mediated protection against lung injury. Increased XLOC_014869 expression promoted apoptosis, and Res inhibited increased levels of this lncRNA, thereby inhibiting LPS-induced lung cell apoptosis. The mechanism by which Res inhibits the increase of XLOC_014869 level is still unclear and should be clarified in future studies.
The proto-oncogene Fos encodes a transcription factor that plays key roles in cell proliferation, differentiation, and apoptosis (36). One study demonstrated that injury can rapidly activate c-Fos in the cerebral cortex (37). c-Fos can be directly transferred to the Fas-L promoter through a single activator protein-1 binding site to induce Fas-L/Fas-mediated apoptosis pathway activation (38). Fas-L expression can also be promoted through the c-Fos/c-Jun and nuclear factor-κB complex (39). Collectively, the results suggest that c-Fos may promote the expression and phosphorylation of c-Jun, thereby promoting Fas-L expression and lung cell apoptosis. These effects were verified with in vitro experiments in rat lung alveolar type II cells with LPS and Res or siRNA.
In conclusion, our findings reveal that lncRNA XLOC_014869 participates the inhibition of Res against LPS-induced lung cell apoptosis, and this may be related to pro-apoptotic factors (c-Fos, c-Jun, and Fas-L). Further research is needed to explore the regulatory mechanism of Res on XLOC_014869, and confirm that inhibiting XLOC_014869 on the protects against ALI.
Acknowledgments
Thanks to Charlesworth Author Services for language polishing.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-1113
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-1113
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-1113). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experiments were approved by the ethics committee of Shanghai Pulmonary Hospital (approval No. K21-025), in compliance with National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Meng L, Li L, Lu S, et al. The protective effect of dexmedetomidine on LPS-induced acute lung injury through the HMGB1-mediated TLR4/NF-κB and PI3K/Akt/mTOR pathways. Mol Immunol 2018;94:7-17. [Crossref] [PubMed]
- Mokra D, Kosutova P. Biomarkers in acute lung injury. Respir Physiol Neurobiol 2015;209:52-8. [Crossref] [PubMed]
- Matthay MA, Zimmerman GA. Acute lung injury and the acute respiratory distress syndrome: four decades of inquiry into pathogenesis and rational management. Am J Respir Cell Mol Biol 2005;33:319-27. [Crossref] [PubMed]
- Villar J, Blanco J, Kacmarek RM. Current incidence and outcome of the acute respiratory distress syndrome. Curr Opin Crit Care 2016;22:1-6. [Crossref] [PubMed]
- Oh YC, Kang OH, Choi JG, et al. Anti-inflammatory effect of resveratrol by inhibition of IL-8 production in LPS-induced THP-1 cells. Am J Chin Med 2009;37:1203-14. [Crossref] [PubMed]
- Sin TK, Yu AP, Yung BY, et al. Modulating effect of SIRT1 activation induced by resveratrol on Foxo1-associated apoptotic signalling in senescent heart. J Physiol 2014;592:2535-48. [Crossref] [PubMed]
- Yang L, Zhang Z, Zhuo Y, et al. Resveratrol alleviates sepsis-induced acute lung injury by suppressing inflammation and apoptosis of alveolar macrophage cells. Am J Transl Res 2018;10:1961-75. [PubMed]
- de Oliveira MTP, de Sá Coutinho D, Tenório de Souza É, et al. Orally delivered resveratrol-loaded lipid-core nanocapsules ameliorate LPS-induced acute lung injury via the ERK and PI3K/Akt pathways. Int J Nanomedicine 2019;14:5215-28. [Crossref] [PubMed]
- Jiang L, Zhang L, Kang K, et al. Resveratrol ameliorates LPS-induced acute lung injury via NLRP3 inflammasome modulation. Biomed Pharmacother 2016;84:130-8. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med 2014;20:623-31. [Crossref] [PubMed]
- Mikawa K, Nishina K, Takao Y, et al. ONO-1714, a nitric oxide synthase inhibitor, attenuates endotoxin-induced acute lung injury in rabbits. Anesth Analg 2003;97:1751-5. [Crossref] [PubMed]
- Wang Q, Zheng X, Cheng Y, et al. Resolvin D1 stimulates alveolar fluid clearance through alveolar epithelial sodium channel, Na,K-ATPase via ALX/cAMP/PI3K pathway in lipopolysaccharide-induced acute lung injury. J Immunol 2014;192:3765-77. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [Crossref] [PubMed]
- Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290-5. [Crossref] [PubMed]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562-78. [Crossref] [PubMed]
- Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007;35:W345-9 [Crossref] [PubMed]
- Sun L, Luo H, Bu D, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 2013;41:e166 [Crossref] [PubMed]
- Finn RD, Mistry J, Schuster-Böckler B, et al. Pfam: clans, web tools and services. Nucleic Acids Res 2006;34:D247-51. [Crossref] [PubMed]
- Li A, Zhang J, Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 2014;15:311. [Crossref] [PubMed]
- Anders S. Analysing RNA-Seq data with the DESeq package 2011. Available online: https://bioc.ism.ac.jp/packages/2.10/bioc/vignettes/DESeq/inst/doc/DESeq.pdf
- Liu ZF, Zheng D, Fan GC, et al. Heat stress prevents lipopolysaccharide-induced apoptosis in pulmonary microvascular endothelial cells by blocking calpain/p38 MAPK signalling. Apoptosis 2016;21:896-904. [Crossref] [PubMed]
- Sureshbabu A, Syed M, Das P, et al. Inhibition of Regulatory-Associated Protein of Mechanistic Target of Rapamycin Prevents Hyperoxia-Induced Lung Injury by Enhancing Autophagy and Reducing Apoptosis in Neonatal Mice. Am J Respir Cell Mol Biol 2016;55:722-35. [Crossref] [PubMed]
- Koh SS, Ooi SC, Lui NM, et al. Effect of Ergothioneine on 7-Ketocholesterol-Induced Endothelial Injury. Neuromolecular Med 2021;23:184-98. [Crossref] [PubMed]
- Alghetaa H, Mohammed A, Sultan M, et al. Resveratrol protects mice against SEB-induced acute lung injury and mortality by miR-193a modulation that targets TGF-β signalling. J Cell Mol Med 2018;22:2644-55. [Crossref] [PubMed]
- Hsieh SC, Lu CC, Horng YT, et al. The bacterial metabolite 2,3-butanediol ameliorates endotoxin-induced acute lung injury in rats. Microbes Infect 2007;9:1402-9. [Crossref] [PubMed]
- Kim G, Piao C, Oh J, et al. Self-assembled polymeric micelles for combined delivery of anti-inflammatory gene and drug to the lungs by inhalation. Nanoscale 2018;10:8503-14. [Crossref] [PubMed]
- Baradaran Rahimi V, Rakhshandeh H, Raucci F, et al. Anti-Inflammatory and Anti-Oxidant Activity of Portulaca oleracea Extract on LPS-Induced Rat Lung Injury. Molecules 2019;24:139. [Crossref] [PubMed]
- Hsieh YH, Deng JS, Chang YS, et al. Ginsenoside Rh2 Ameliorates Lipopolysaccharide-Induced Acute Lung Injury by Regulating the TLR4/PI3K/Akt/mTOR, Raf-1/MEK/ERK, and Keap1/Nrf2/HO-1 Signaling Pathways in Mice. Nutrients 2018;10:1208. [PubMed]
- Wang J, Shen YC, Chen ZN, et al. Microarray profiling of lung long non-coding RNAs and mRNAs in lipopolysaccharide-induced acute lung injury mouse model. Biosci Rep 2019;39:BSR20181634 [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Zhou Z, Zhu Y, Gao G, et al. Long noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia. Life Sci 2019;228:189-97. [Crossref] [PubMed]
- Li H, Shi H, Gao M, et al. Long non-coding RNA CASC2 improved acute lung injury by regulating miR-144-3p/AQP1 axis to reduce lung epithelial cell apoptosis. Cell Biosci 2018;8:15. [Crossref] [PubMed]
- Wang L, Liu J, Xie W, et al. Overexpression of MALAT1 Relates to Lung Injury through Sponging miR-425 and Promoting Cell Apoptosis during ARDS. Can Respir J 2019;2019:1871394 [Crossref] [PubMed]
- Habib GM. Arsenite causes down-regulation of Akt and c-Fos, cell cycle dysfunction and apoptosis in glutathione-deficient cells. J Cell Biochem 2010;110:363-71. [Crossref] [PubMed]
- Nagy Z, Simon L, Bori Z. Regulatory mechanisms in focal cerebral ischemia. New possibilities in neuroprotective therapy. Ideggyogy Sz 2002;55:73-85. [PubMed]
- Chen X, Shen J, Wang Y, et al. Up-regulation of c-Fos associated with neuronal apoptosis following intracerebral hemorrhage. Cell Mol Neurobiol 2015;35:363-76. [Crossref] [PubMed]
- Kasibhatla S, Brunner T, Genestier L, et al. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-kappa B and AP-1. Mol Cell 1998;1:543-51. [Crossref] [PubMed]


