
Whole exome sequencing identifies novel inherited genetic variants in tetralogy of Fallot
Introduction
Tetralogy of Fallot (TOF) is a combined congenital heart malformation characterized by the simultaneous presence of pulmonary artery stenosis, aortic straddle, ventricular septal defect, and right ventricular hypertrophy, resulting in hemodynamic changes such as right ventricular hypertension and hypertrophy (1). TOF is one of the most common cyanotic congenital heart diseases (CHDs). It affects about 3–5 per 10,000 newborns and represents 7%-10% of all CHD cases (2,3). With dramatic advances in surgery and medication, the early survival of TOF patients has improved significantly, but long-term sequelae, including cardiac dysfunction and arrhythmia, still cause great distress to most TOF patients (4). A better understanding of the possible causes of TOF will help us better understand the disease’s pathophysiology and help reduce the risk of disease development.
TOF is closely related to prenatal infection, teratogenic exposure, maternal disease, and genetic factors, and rare genetic variants have been confirmed as important risk factors of TOF. Previous studies have identified that a number of genetic variants, such as NKX2-5, GATA4, TBX5, ZIC3, FOXH1, NODAL, and GJA1, are involved in TOF and other cardiac defects (5,6). A study on exome sequencing of 2,871 CHD patients confirmed the important contribution of GDF1, MYH6, and FLT4 mutations in the pathogenesis of CHD (7). By analyzing exome sequencing data from 811 probands with TOF, Reuter et al. (8) identified likely causative variants in FLT4 and NOTCH1, and revealed 1–3 variants in 21 other genes, including ATRX, DLL4, EP300, GATA6, JAG1, NF1, PIK3CA, RAF1, RASA1, SMAD2, and TBX1. Manshaei et al. (9) confirmed the involvement of FLT4 truncating variants and NOTCH1 missense variants in TOF, accounting for 11–14% of individuals in the TOF cohort. Page et al. (10) assessed the genetic variants in 829 non-syndromic TOF patients and confirmed that the NOTCH1 gene variants are the most frequent genetic variants in non-syndromic TOF, followed by FLT4, accounting for almost 7% of TOF patients. Lin and his colleagues identified mutations in PEX5, NACA, ATXN2, CELA1, PCDHB4 and CTBP1 as potential genetic risk factors of sporadic TOF (11).
Although changes in genetic material associated with TOF have been reported more frequently, there have been fewer reports of simple and sporadic TOF. Familial studies have shown that 80% of patients with sporadic CHD may have significant, complex genetic conditions or single nucleotide polymorphisms (SNPs), while 20% of the remaining CHD patients have chromosomal abnormalities or syndromes of multi-system malformation. Although genetic studies using next-generation sequencing technology have revealed the involvement of hundreds of genetic variants in TOF, these are not sufficient to fully elucidate the pathology of TOF.
In this study, we examined the genetic information of 19 sporadic TOF patients by whole exome sequencing (WES) and screened possible pathological variations by bioinformatics analysis. In addition, Sanger sequencing was used to verify these pathological mutations. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-970/rc).
Methods
Study design and participants
A cohort of 19 unrelated patients who received surgical TOF treatment were recruited from Guizhou Provincial People’s Hospital between March 2018 and July 2021 in this study. TOF was confirmed by echocardiography, clinical symptoms, signs, and intraoperative findings. The detailed phenotyping data was listed in Table 1. The 19 patients aged 9 months to 34 years included 8 females and 11 males. This study was a non-randomized and double-blind trial. The study protocol was approved by the Ethics Committee of Guizhou Provincial People’s Hospital (No. 2018040) and performed in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from enrolled participants or participants’ parents.
Table 1
| Subjects | Age (years) | Sex | RV diastolic diameter (mm) | RVAW (thickened) (mm) |
RVOT (mm) | Acropachia | Cyanosis |
|---|---|---|---|---|---|---|---|
| TOF-1 | 16 | F | 15.1 | – | 7.6 | Yes | Yes |
| TOF-2 | 11 | M | 15 | – | 7.4 | Yes | Yes |
| TOF-3 | 0.83 | M | 15.5 | – | 6 | No | Yes |
| TOF-4 | 0.75 | F | 8.5 | – | 7 | No | Yes |
| TOF-5 | 1 | M | 18.6 | 6.9 | – | Yes | Yes |
| TOF-6 | 2 | M | 10.3 | 5.7 | 5.4 | Yes | Yes |
| TOF-7 | 24 | F | 21 | 8.6 | – | Yes | Yes |
| TOF-8 | 22 | M | 16.1 | 7.0 | – | No | No |
| TOF-9 | 34 | F | 30 | 11 | 7.7 | No | No |
| TOF-10 | 1 | F | 13.4 | 6.4 | – | No | Yes |
| TOF-11 | 1 | M | 10 | 9.3 | – | No | Yes |
| TOF-12 | 5 | M | 11.1 | 5.3 | 4.8 | No | Yes |
| TOF-13 | 13 | F | 20.7 | 10.9 | – | Yes | Yes |
| TOF-14 | 2 | F | 12.3 | 5.4 | 8.9 | No | Yes |
| TOF-15 | 4 | F | – | – | – | No | No |
| TOF-16 | 2 | F | 10.4 | 8.1 | – | No | Yes |
| TOF-17 | 1 | F | 13.8 | 6.1 | 5 | Yes | Yes |
| TOF-18 | 1 | M | 9.6 | 4.6 | 7.5 | No | No |
| TOF-19 | 2 | F | 12.3 | 6.8 | 5.9 | Yes | Yes |
TOF, tetralogy of Fallot; F, female; M, male; RV, right ventricle; RVAW, right ventricular anterior wall; RVOT, right ventricular outflow tract.
DNA extraction
For DNA testing, 10 mL of peripheral blood was collected in an EDTA-containing tube. The isolation of genomic DNA from the peripheral blood of the patient was performed with the EasyPure Blood Genomic DNA kit (TransGen Biotech, Beijing, China) according to the standard operating protocol. The quality and quantity of DNA samples were analyzed using a NanoDrop spectrophotometer (Thermo Fisher, USA).
WES analysis
WES analysis was performed on 19 patients with sporadic TOF at ANOROAD (Beijing, China). In brief, the sequencing library was prepared using the SureSelectXT Target Enrichment kit (Agilent, Santa Clara, CA, USA) and captured using the Agilent SureSelect Human Whole Exon kit V5 (Agilent, Santa Clara, CA, USA). Double-terminal sequencing was performed using the HiSeq2500PE100 platform (Illumina, San Diego, CA, USA). The reading length of each sample was 100 bp, and the average coverage depth was at least 100×. Each sample was repeated independently three times.
Data analysis
Base calling and quality control were conducted by real-time analysis on the NextSeq500 system. The BCL files were converted into FASTQ files using Bcl2fastq Conversion Software. The whole sequenced data were trimmed for low-quality sequences and aligned to UCSC human reference genome (GRCh38/HG38) using Burrows-Wheeler Alignment (BWA). The Genome Analysis Toolkit (GATK) and VarScan were used to detect SNPs and small insertions/deletions. ANNOVAR was used to annotate the variants with several databases including dbSNP, GnomAD (12), 1000 Genomes Project (13), and ExAC (14). Finally, four online mutation pathogenicity prediction and analysis software including PolyPhen, SIFT, MutationTaster, and FATHMM were used to predict the influence of polymorphic variation on coding proteins and conservation, so as to conduct mutation pathogenicity analysis.
Variant validation
Sanger sequencing was used to validate candidate variants from WES. The sequences of primers for PCR amplification are shown in Table 2. The ABI PRISM BigDye kit and ABI 3130 DNA sequencer (Applied Biosystems, Carlsbad, USA) were used for sequencing. Sequencing data were analyzed with Chromas software (version 2.23). The samples were repeated independently three times
Table 2
| Genes | Primer sequence |
|---|---|
| RNF135 | Forward: 5' GCTGGAGCTGTGAGAGGTTT 3' |
| Reverse: 5' CAGGTCTGTCTGAGCCAAGG 3' | |
| APOB | Forward: 5' AAGGGTTCGGTTCTTTCTCGG 3' |
| Reverse: 5' AGAGAGTTCCAGGGTGGCTT 3' |
PCR, polymerase chain reaction.
Results
A total of 19 TOF patients and 3 healthy volunteers were included in this study. The clinical characteristics of the participants were shown in Table 1. For all subjects, the age at diagnosis ranged from 9 months to 34 years.
WES analysis was performed on DNA samples from 19 TOF patients and 3 healthy controls. Considering that the incidence of TOF is 1 in 3000 live births, variants with a minor allele frequency (MAF) of less than 1% were retained. Then, only missense mutations, frame-shift mutations, nonsense mutations, and intron splicing site mutations were retained. As a result, 21 genetic variants involving 16 genes were found in 12 patients with sporadic TOF (Table 3). The types of mutations included missense and splicing variants. None of these genes were detected in samples from the 3 healthy controls.
Table 3
| Subjects | Gene | Nucleotide variation | Amino acid variation | Pathogenicity | Frequency (f) |
|---|---|---|---|---|---|
| 1 | Not detected | ||||
| 2 | TBX1 | NM_001379200.1:c.1001C>T | p.Thr334Met | LiPath | NA |
| 3 | CD96 | NM_001318889.2:c.791C>T | p.Thr264Met | Path | 8.2e-6 |
| 4 | BRCA1 | NM_007294.4:c.3257T>A | p.Leu1086Ter | Path | NA |
| RNF135 | NM_032322.4:c.1015del | p.Val339fs | Path | 1e-4 | |
| TBX1 | NM_005992.1:c.929G>C | p.Gly310Ala | VUS | 3.5e-5 | |
| 5 | Not detected | ||||
| 6 | Not detected | ||||
| 7 | G6PD | NM_001360016.2:c.1388G>A | p.Arg463His | Path | |
| 8 | ABCC6 | NM_001171.5:c.232G>A | p.Ala78Thr | Path | NA |
| 9 | Not detected | ||||
| 10 | Not detected | ||||
| 11 | NF1 | NM_001042492.2:c.3198-2A>T | Splicing | LiPath | NA |
| 12 | KCNQ4 | NM_004700.4:c.546C>G | p.Phe182Leu | Path | 3e-4 |
| APOB | NM_000384.3:c.10700C>T | p.Thr3567Met | LiPath | 7.4e-5 | |
| 13 | PNPLA2 | NM_020376.4:c.757+1G>T | Splicing | Path | NA |
| NF1 | NM_001042492.2:c.3198-2A>T | Splicing | LiPath | NA | |
| 14 | Not detected | ||||
| 15 | KLF13 | NM_001302461.2:c.319T>A | p.Ser107Thr | VUS | NA |
| 16 | KLF13 | NM_001302461.2:c.310G>C | p.Glu104Gln | VUS | NA |
| TBX15 | NM_001330677.2:c.980G>A | p.Arg327His | VUS | 5e-4 | |
| ROM1 | NM_000327.3:c.339dupG | p.Leu114fs | LiPath | 1.6e-5 | |
| 17 | FLG | NM_002016.1:c.7264G>T | p.Glu2422Ter | Path | 1.9e-4 |
| NF1 | NM_001042492.2:c.3198-2A>T | Splicing | LiPath | NA | |
| KLF13 | NM_001302461.2:c.319T>A | p.Ser107Thr | VUS | NA | |
| 18 | GATA4 | NM_002052.3:c.191G>A | p.Gly64Glu | Path (VSD) | NA |
| FOXC2 | NM_005251.2:c.794A>G | p.Asn265Ser | VUS | NA | |
| 19 | Not detected | ||||
TOF, tetralogy of Fallot; VUS, variants of unknown significance; VSD, ventricular septal defect; NA, not available.
By reviewing exome sequencing databases including the dbSNP, GnomAD, 1000 Genomes and ExAC, 13 of the 21 variants identified in this study had allele frequencies of 0 in these databases, indicating that these variants are very rare. Then, 4 online pathogenicity prediction and analysis software, including Polyphen-2, SIFT, MutationTaster, and FATHMM, were used to analyze the variant pathogenicity at the bioinformatics level. We found 9 pathogenic variants, 6 variants that might be dangerous, and 6 variants that were not dangerous (Table 3).
Furthermore, we analyzed the clinical symptoms of TOF patients with gene variations and found that patients with APOB and RN135 variants had more serious clinical symptoms. The 2 variations were then analyzed using Sanger sequencing, and the results showed that the 2 variants were heterozygous in the patients (Figure 1).
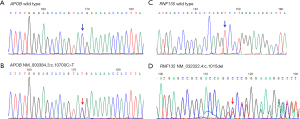
Discussion
In this study, we performed WES on DNA samples from 19 patients with TOF. We identified 21 variants related to TOF that were found in 12 patients, including 9 pathogenic variants, 6 suspected pathogenic variants, and 6 variants of unknown significance (VUS). Patients who had heterozygous APOB and RNF135 variants had more severe symptoms of TOF, which indicates that they may be important genetic factors for sporadic cases of TOF as well.
WES, the high-throughput sequencing of whole genome exon regions by sequence capture method, has been used to investigate coding variation. The exome of 30 million bp represents about 1% of the human genome, but accounts for about 80% of disease-related variation (15). Therefore, WES is a cost-effective method for TOF-related variants. Previous studies have revealed several rare variants of TOF via WES technology. Wang et al. (16) revealed a novel missense variant of MYOM2 associated with TOF by analyzing WES data from a Chinese family whose twins were affected by TOF. Several WES analyses for fetuses with antenatal diagnosis of TOF identified de novo heterozygous frameshift variants in SMARCC2 and one homozygous variant in OTUD6B (17,18). In this study, WES was used to characterize the genetic information of 19 TOF patients and 3 healthy controls. The reads mapping and variant calling of WES data was performed as previously reported (19), and the variants were filtered and annotated following the standards and guidelines for the interpretation of sequence variants (20). Finally, we identified 21 genetic variants involving 16 genes in 12 patients with sporadic TOF. Importantly, we found that APOB and RN135 were associated with serious clinical symptoms of TOF.
The APOB protein is the major apolipoprotein that carries chylomicron and low density lipoprotein (LDL). There are two isoforms of APOB in plasma, namely APOB-48 and APOB-100. The two isoforms have the same N-terminal sequence. The shorter APOB-48 protein was tested against residue 2180. The APOB-100 transcript is produced after RNA editing, resulting in the termination codon and premature termination of translation. Mutations in APOB gene result in low lipoproteinemia, normal triglyceridemia, and hypercholesterolemia due to ligand-deficient APOB, as well as disorders that affect plasma cholesterol and APOB levels (21,22). APOB has been identified as a causative gene of familial hypercholesterolemia (FH) (23,24). The investigation from Benedek et al. (25) showed that APOB c.10580G>A is the most common mutation in Swedish patients with FH. The agnostic genetic investigation by Zuber et al. (26) prioritized APOB as a key lipid risk factor for coronary artery diseases. In our study, we found that the TOF patient with APOB c.10700C>T presented with severe developmental defects of the heart, indicating the possible pathogenic activity of the APOB variant.
The RNF135 gene is located at the NF1 locus of 17q11.2, encoding a protein containing a RING finger domain at the N-terminal. The RING domain is a zinc finger domain with ubiquitin and sumo ligase activity. Several studies have indicated the correlation between RNF135 mutations and neuronal diseases. The RNF135 gene is located in a chromosomal region that is often frequently absent in patients with neurofibromatosis (27,28). Furthermore, Tastet et al. (29) showed a significant increase in the frequency of genotypes carrying a missense variant of the rare allele rs111902263 (p.R115K) in a cohort of French patients with autism, while three unrelated patients showed a homozygous genotype for K115. Besides, mutations in the RNF135 gene were also associated with an overgrowth syndrome (30). So far, RNF135 has not been reported to be associated with cardiovascular disease. In our study, we found that RNF135 c.1015del was observed in patients with TOF. Considering the extensive function and importance of this gene in protein-protein and protein-DNA interactions, we speculated that the RNF135 mutation might be a novel pathogenic mutation in TOF.
However, there are still some problems to be solved before these possible pathogenic genes can be used as diagnostic markers or therapeutic targets for TOF. Firstly, the relationship between variants in APOB or RNF135 and TOF needs to be analyzed on large cohorts. Furthermore, the effects of these variants on protein structure, stability and expression need to be uncovered. In addition, the potential functional involvement of these genes in the pathogenesis of TOF needs to be explored by further in vivo and in vitro experiments.
In this study, we identified several genetic variants associated with TOF and confirmed that variants of RNF135 and ABOB were associated with TOF severity. These findings contribute to the genetic etiopathogenesis of TOF.
Acknowledgments
Funding: This study was supported by the Guizhou Science and Technology Support Program (Guizhou Science and Technology Cooperation Support 2018-2783) and the National Natural Science Foundation of China (No. 81860273).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-970/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-970/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-970/coif). YP reports that this study was supported by the Guizhou Science and Technology Support Program (Guizhou Science and Technology Cooperation Support 2018-2783) and the National Natural Science Foundation of China (No. 81860273). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Ethics Committee of Guizhou Provincial People’s Hospital (No. 2018040) and performed in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from enrolled participants or participants’ parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Blais S, Marelli A, Vanasse A, et al. The 30-Year Outcomes of Tetralogy of Fallot According to Native Anatomy and Genetic Conditions. Can J Cardiol 2021;37:877-86. [Crossref] [PubMed]
- van der Linde D, Konings EE, Slager MA, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol 2011;58:2241-7. [Crossref] [PubMed]
- Zhou Y, Mao X, Zhou H, et al. Birth Defects Data From Population-Based Birth Defects Surveillance System in a District of Southern Jiangsu, China, 2014-2018. Front Public Health 2020;8:378. [Crossref] [PubMed]
- Nathan M, Levine JC, Van Rompay MI, et al. Impact of Major Residual Lesions on Outcomes After Surgery for Congenital Heart Disease. J Am Coll Cardiol 2021;77:2382-94. [Crossref] [PubMed]
- Su W, Zhu P, Wang R, et al. Congenital heart diseases and their association with the variant distribution features on susceptibility genes. Clin Genet 2017;91:349-54. [Crossref] [PubMed]
- Kalayinia S, Maleki M, Mahdavi M, et al. Whole-Exome Sequencing Reveals a Novel Mutation of FLNA Gene in an Iranian Family with Nonsyndromic Tetralogy of Fallot. Lab Med 2021;52:614-8. [Crossref] [PubMed]
- Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet 2017;49:1593-601. [Crossref] [PubMed]
- Reuter MS, Chaturvedi RR, Jobling RK, et al. Clinical Genetic Risk Variants Inform a Functional Protein Interaction Network for Tetralogy of Fallot. Circ Genom Precis Med 2021;14:e003410. [Crossref] [PubMed]
- Manshaei R, Merico D, Reuter MS, et al. Genes and Pathways Implicated in Tetralogy of Fallot Revealed by Ultra-Rare Variant Burden Analysis in 231 Genome Sequences. Front Genet 2020;11:957. [Crossref] [PubMed]
- Page DJ, Miossec MJ, Williams SG, et al. Whole Exome Sequencing Reveals the Major Genetic Contributors to Nonsyndromic Tetralogy of Fallot. Circ Res 2019;124:553-63. [Crossref] [PubMed]
- Liu L, Wang HD, Cui CY, et al. Whole exome sequencing identifies novel mutation in eight Chinese children with isolated tetralogy of Fallot. Oncotarget 2017;8:106976-88. [Crossref] [PubMed]
- Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434-43. [Crossref] [PubMed]
- 1000 Genomes Project Consortium; Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68-74.
- Karczewski KJ, Weisburd B, Thomas B, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 2017;45:D840-5. [Crossref] [PubMed]
- Wang Z, Liu X, Yang BZ, et al. The role and challenges of exome sequencing in studies of human diseases. Front Genet 2013;4:160. [Crossref] [PubMed]
- Wang J, Wang C, Xie H, et al. Case Report: Tetralogy of Fallot in a Chinese Family Caused by a Novel Missense Variant of MYOM2. Front Cardiovasc Med 2022;9:863650. [Crossref] [PubMed]
- Börklü E, Altunoğlu U, Eraslan S, et al. A New Family with a Novel OTUD6B Mutation: Practicing Whole Exome Sequencing for Antenatal Diagnosis of Tetralogy of Fallot. Mol Syndromol 2022;13:206-11. [Crossref] [PubMed]
- Sun H, Zhang S, Wang J, et al. Expanding the phenotype associated with SMARCC2 variants: a fetus with tetralogy of Fallot. BMC Med Genomics 2022;15:40. [Crossref] [PubMed]
- Shi X, Zhang L, Bai K, et al. Identification of rare variants in novel candidate genes in pulmonary atresia patients by next generation sequencing. Comput Struct Biotechnol J 2020;18:381-92. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Futema M, Ramaswami U, Tichy L, et al. Comparison of the mutation spectrum and association with pre and post treatment lipid measures of children with heterozygous familial hypercholesterolaemia (FH) from eight European countries. Atherosclerosis 2021;319:108-17. [Crossref] [PubMed]
- Futema M, Taylor-Beadling A, Williams M, Humphries SE. Genetic testing for Familial Hypercholesterolaemia - Past, Present and Future. Journal of lipid research. 2021;62:100139. [Crossref] [PubMed]
- Nagahara K, Nishibukuro T, Ogiwara Y, et al. Genetic Analysis of Japanese Children Clinically Diagnosed with Familial Hypercholesterolemia. J Atheroscler Thromb 2022;29:667-77. [Crossref] [PubMed]
- Yang S, Ke X, Liang H, et al. Case Report: A Clinical and Genetic Analysis of Childhood Growth Hormone Deficiency With Familial Hypercholesterolemia. Front Endocrinol (Lausanne) 2021;12:691490. [Crossref] [PubMed]
- Benedek P, Jiao H, Duvefelt K, et al. Founder effects facilitate the use of a genotyping-based approach to molecular diagnosis in Swedish patients with familial hypercholesterolaemia. J Intern Med 2021;290:404-15. [Crossref] [PubMed]
- Zuber V, Gill D, Ala-Korpela M, et al. High-throughput multivariable Mendelian randomization analysis prioritizes apolipoprotein B as key lipid risk factor for coronary artery disease. Int J Epidemiol 2021;50:893-901. [Crossref] [PubMed]
- Brussa Reis L, Turchetto-Zolet AC, Fonini M, et al. The Role of Co-Deleted Genes in Neurofibromatosis Type 1 Microdeletions: An Evolutive Approach. Genes (Basel) 2019;10:839. [Crossref] [PubMed]
- Rosset C, Vairo F, Cristina Bandeira I, et al. Clinical and molecular characterization of neurofibromatosis in southern Brazil. Expert Rev Mol Diagn 2018;18:577-86. [Crossref] [PubMed]
- Tastet J, Decalonne L, Marouillat S, et al. Mutation screening of the ubiquitin ligase gene RNF135 in French patients with autism. Psychiatr Genet 2015;25:263-7. [Crossref] [PubMed]
- Douglas J, Cilliers D, Coleman K, et al. Mutations in RNF135, a gene within the NF1 microdeletion region, cause phenotypic abnormalities including overgrowth. Nat Genet 2007;39:963-5. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


