
Screening of sphingolipid metabolism-related genes associated with immune cells in myocardial infarction: a bioinformatics analysis
Introduction
Myocardial infarction (MI) is a cardiovascular event caused by prolonged severe myocardial ischemia and ischemic necrosis due to a sharp reduction or interruption of coronary blood supply (1). The main clinical presentation of MI is typical ischemic chest discomfort, which can be accompanied by nausea, vomiting, sweating, dyspnea, and/or syncope (2). In the United States, the overall prevalence of MI among adults over the age of 20 is 3%. Every 40 s about 1 American develops MI. MI is also one of the leading causes of mortality worldwide. With a 5-year survival rate of approximately 30%, MI has a tremendous impact on health and the global economy (3). The degree of MI is positively associated with the risk of developing heart failure after infarction, during which ventricular remodeling is a prominent pathophysiological process (4). The immune system is particularly important after MI (5). The pro-inflammatory response triggered by MI can clear necrotic cells and debris; if over-activated, however, it causes severe damage to the extracellular matrix (6). Therefore, the inflammatory response plays a dual role in MI prognosis. Maintaining the balance of the inflammatory response is crucial for improving cardiac prognosis and protecting ventricular function. Activation of various immune cells (e.g., neutrophils, lymphocytes, monocytes, and macrophages) and their infiltration into tissues after MI have been demonstrated (7), but are still under investigation. Therefore, targeting the immune system and resolving inflammation may be a therapeutic option for cardioprotection after MI.
The immune system is involved in complex physiological processes. Serving as a keeper of dynamic balance (8,9), it senses and integrates environmental signals by recruiting various immune cells and responds to different pathological circumstances, thus ensuring cell viability and long-term survival. The immune response to various challenges results in dramatic changes involving numerous signaling molecules and intricate cell-to-cell interactions (10). Sphingolipid metabolites (SMs), in particular ceramide (N-acyl-sphingosine), ceramide-1-phosphate (C1P), and sphingosine-1-phosphate (S1P), are bioactive lipid molecules that have been shown to regulate a wide variety of cellular processes and play key roles in immunity, inflammation, and inflammatory diseases (11). Composed of ceramides and various types of glycolipids, SMs are ubiquitous components in eukaryotic membranes. They serve as an important second messenger in immune cells and play key roles in cellular signal transduction. S1P, a key mediator of SM, is involved in regulating cell proliferation, differentiation, and survival. Ceramide is a key precursor to bioactive SMs and transmits intracellular signals during cell cycle arrest, apoptosis, autophagy, and death (12). While some targets of sphingolipid signaling have been identified, how it modulates immune responses remains unknown. A large body of evidence suggests that SM plays an important regulatory role in MI. Blood ceramide level can be marked as a predictor of the severity of cardiovascular complications and mortality (13). It was found that the S1P/ceramide ratio in the non-infarcted area of the left ventricle of MI rats was significantly decreased, suggesting that the early activation of apoptosis in the non-infarcted area of the left ventricle after MI may be related to a decreased ratio (14). Pharmacological elevation of bioactive lipids after acute MI leads to improvements in cardiac structure and function (15). Hadas et al. (16) suggested that acid ceramidase attenuated detrimental neutrophil levels and cell death in the left ventricle after MI by altering SM, and inhibition of ceramide de novo synthesis could reduce myocardial reperfusion injury and inflammation (17). In a lipidomic study, the expression of ceramide kinase, which phosphorylates ceramide to produce C1P, was found to be consistently upregulated in myocardial tissue after MI (18).
In our current study, we utilized bioinformatics technology to investigate MI-related genes and immune mechanisms, provided further insights into the roles of SMs in the immune system and their interactions, and attempted to explain the possible roles of SM-related genes in disease progression after MI. We present the following article in accordance with the STREGA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1041/rc).
Methods
Identification of differentially-expressed genes (DEGs) in the Gene Expression Omnibus (GEO) dataset
GSE61145 is a human peripheral blood dataset and GSE23294 and GSE71906 are 2 mouse myocardial tissue datasets. All datasets were selected from the GEO database. GSE61145 includes 7 normal myocardial tissues and 17 MI specimens, whereas GSE23294 and GSE71906 contain 10 normal myocardial tissues and 10 MI specimens. We downloaded the platform and matrix files and processed them using the R package and annotation packages. The probe names were converted to the international standard names (gene symbols) for the genes. MI and normal samples from 3 microarray datasets were screened using BioConductor (http://www.bioconductor.org/), setting the |fold change (FC)| as ≥0.75 and the adjusted P value cutoff as <0.05 for the screening of DEGs. The 3 DEG datasets were integrated by batch normalization in R. We obtained upregulated and downregulated genes in these 3 profiles for subsequent analyses. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses
To gain a deeper understanding of the selected DEGs and key modules, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (19) was used for the GO and KEGG enrichment analyses (20), during which a pathway database containing information on how molecules or genes are networked was applied, with the adjusted P value cutoff being <0.05. Immune network pathways were identified using the Cytoscape plugin ClueGO.
Assessment of immune cell infiltration
The gene expression levels in the integrated dataset were normalized using a format acceptable to “Cell type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT)” and then the data were uploaded to the CIBERSORT web portal (http://cibersort.stanford.edu/), which quantifies cell composition from bulk gene expression profiles (GEPs) (21). The types of infiltrating immune cells were predicted using the CIBERSORT algorithm (P value <0.05). To analyze the significant differences in immune cell expression levels among different cell types,
Verification of SM-related gene expression in neutrophils in the single-cell sequencing database GSE130699
The MI mouse single-cardiomyocyte RNA sequencing dataset (GSE130699) was downloaded from the GEO database. A total of 2 normal and 2 MI samples were included. MI was induced in neonatal mice on the 1st or 8th day after birth by permanent ligation of the anterior descending coronary artery, and myocardial tissue was collected on the 1st and 3rd days after the operation. The surgery was regarded as successful after TTC staining confirmed the death of myocardial cells. After annotation, the cell types were identified in the normal group and the modeling group.
Statistical analysis
Data were analyzed using SPSS 22.0 software. The differences in SM-related gene expression levels in neutrophils were compared using the Wilcoxon rank sum test. P<0.05 was considered to represent statistical significance.
Results
Identification of 101 immune-related DEGs
To gain a comprehensive understanding of MI genomic expression profiles, the MI expression profile dataset (GSE61145) was normalized, and a total of 181 upregulated and 80 downregulated genes were screened (Table S1). Figure 1A is the volcano plot of DEGs in this dataset, in which 101 immune-related DEGs were identified. Immune genes were significantly enriched in biological processes including neutrophil migration, T cell activation, and T cell differentiation (Figure 1B). After screening by CIBERSORT, 7 normal peripheral blood samples and 17 MI peripheral blood samples were obtained. Three types of immune cells were identified, namely neutrophils, resting memory CD4+ T cells, and γδ T cells (Figure 1C).
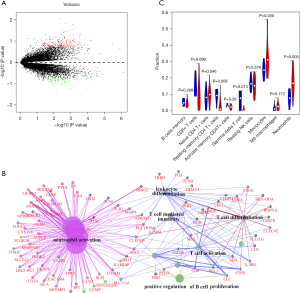
GO functional and KEGG pathway enrichment analysis of 185 upregulated genes and 9 downregulated genes
To gain insight into MI genomic expression profiles, 2 additional mouse MI expression profile datasets (GSE23294 and GSE71906) were normalized, and a total of 185 upregulated and 9 downregulated genes were screened (Table S2). Figure 2A is a volcano plot for these 2 datasets. In addition, a heatmap was used to plot the top 15 upregulated and downregulated genes, as shown in Figure 2B. These genes included Sprr1a, Adamts4, Fstl3, Hmox1, Cdr2, Empl, Anxa2, Ctgf, Col4a1, Tpm4, Timp1, Uck2, Rcan1, Tubb6, and Actn1.
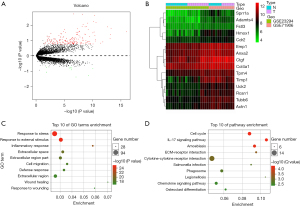
To further understand these 2 mouse DEGs datasets, we performed GO functional and KEGG pathway enrichment analyses for DEGs using the DAVID database. Different genes were particularly abundant in biological processes including response to stress, response to external stimulus, inflammatory response, extracellular space, extracellular region part, cell migration, defense response, extracellular region, wound healing, and response to wounding (Figure 2C). Pathways involving different genes are shown in Figure 2D, including the cell cycle, IL-17 signaling pathway, amoebiasis, ECM-receptor interaction, cytokine-cytokine receptor interaction, Salmonella infection, phagosome, legionellosis, chemokine signaling pathway, and osteoclast differentiation.
Expression of SM-related genes in myocardial tissues
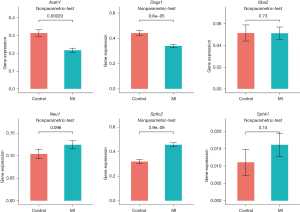
We analyzed the differences in SM-related genes obtained from GSE23294 and GSE71906 in myocardial tissues of MI mice. Among them, the expression levels of Asah1, Degs1, Neu1, Sptlc2, and Sphk1 were significantly upregulated, and the expression level of Gba2 was downregulated (Figure 3).
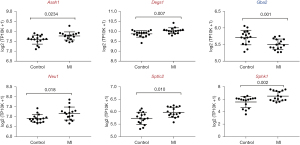
Differential expression of SM-related genes in neutrophils
Identification of DEGs from expression profiles by microarrays (GSE611145, GSE23294, and GSE71906) reflects the average differential expression of genes in multiple cells, whereas single-cell sequencing can reflect the genomic and transcriptomic statuses of individual cells. Furthermore, we analyzed the differential expression of SM-related genes screened in myocardial tissue obtained from GSE130699. After annotation, 12 distinct cell populations were identified from the single-cell sequencing data (Figure 4). A total of 3,896 neutrophils were recorded. The normal group included 657 neutrophils and the MI group included 3239 neutrophils. The expression levels of Asah1, Degs1, and Sptlc2 were significantly different in neutrophils (Figure 5). Notably, the expression of Sptlc2 was significantly upregulated in neutrophils of the MI group, while the expression levels of Asah1 and Degs1 were downregulated.
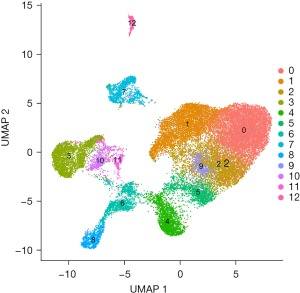
Discussion
A variety of immune cells, inflammatory cytokines, enzymes, lipid mediators, and cellular molecules participate in the occurrence and development of MI via complex mechanisms. Our data showed that some genes were significantly enriched in immune regulation. Subsequent analyses also revealed that neutrophils and T cells are involved in MI. Among immune cells, neutrophils, which are a type of polymorphonuclear leukocyte, are considered as a major player in the innate immune system and are also the first leukocytes recruited to the sites of acute inflammation for clearing infection (22,23). In another study, however, neutrophils played further roles in chronic inflammatory conditions and in adaptive immune responses (24). A study in SM has shown that both ceramides and S1P are involved in the regulation of neutrophil recruitment, phagocytosis, and migration (25). As pro-apoptotic metabolites, they also mediate neutrophil apoptosis through caspase activation (26). T cells are another type of immune cell. As the main trigger of many types of inflammation, they consist of multiple subpopulations that play different roles at different life stages and have the potential to recognize antigens, maintain immune memory, and develop self-tolerance (27). A study has found that acid sphingomyelinase-dependent ceramide signaling plays a key role in mediating the activation, proliferation, and response of CD4+ T cells (28). Furthermore, a study has shown that targeting macrophages can prevent left ventricular remodeling and physiological deterioration after MI (29); however, no prominent changes in macrophages were found in our current study. One possible explanation is that the types of immune cells were simply the expression patterns inferred from genes.
Metabolic characterization of the myocardium suggests that cardiometabolic disturbances underlie most cardiovascular diseases. Untargeted mass spectrometry-based analysis found that changes in sphingomyelin metabolism were associated with cardiovascular diseases (30), suggesting that both immune and metabolic disorders are involved in the occurrence and development of MI. The link between metabolism and immune responses is not limited to MI but also has potential roles in other diseases. Sphingolipid signaling is involved in regulating the functions of immune cells including neutrophils, macrophages, NK cells, CD8+ T cells, and CD4+ T cells. Recent evidence suggests that activation of the S1P pathway inhibits neutrophil apoptosis in acute lung injury (31). In contrast, neutrophil elastase has also been reported to increase neutrophil counts and airway ceramide levels in lung disease (32). One of the main metabolic pathways in MI is SM. A previous study has proposed that S1P-mediated signaling changes in the heart after MI have the in vitro potential to enhance cardiomyocyte survival and provide cardioprotection. Furthermore, SMs released by myocardial tissue during ischemic stress or inflammation may become therapeutic targets for cardiovascular diseases (33). It has been found that SM can be altered after MI by hydrolyzing ceramides in the heart, thereby supporting cardiac function by limiting neutrophil infiltration and altering the immune response (16).
In the identification of human DEGs, we found from CIBERSORT and ClueGO that immune genes were significantly enriched in biological processes such as neutrophil migration and T cell activation/differentiation. Literature review and analyses of single-cell sequencing datasets further showed a correlation between neutrophils and SM. Therefore, in our current study, we explored the relationship between neutrophils and SM in MI. Six DEGs were found to be highly correlated with SM. Among them, Asah1, Degs1, and Sptlc2 were significantly differentially expressed in neutrophils. Some enzymes are the products of these DEGs and can functionally achieve precise regulation of SMs and neutrophils.
Acid ceramidase (Asah1) is a key mature heterodimeric enzyme that regulates intracellular ceramide metabolism by catalyzing the hydrolysis of ceramides to sphingosine and fatty acids (34). Ceramides and sphingosines are important interconvertible SM metabolites that control multiple signaling pathways related to different aspects of cell survival and aging (35). It has been shown that high-mobility group box-1 (HMGB1) may damage the vasculature system through inhibition of Asah1 and subsequent accumulation of long-chain ceramides in coronary cardiomyocytes (36). Ceramides are a class of lipids thought to be toxic, and their pathway enzymes are upregulated in ischemic myocardium, which may be one of the sources of plasma ceramide during MI (37). Furthermore, S1P generated by Asah1 phosphorylation has been shown to inhibit inflammatory neutrophil recruitment and cardiomyocyte apoptosis and may stimulate tissue regeneration and improve cardiac function by attracting hematopoietic stem cells to the infarct site (38). The release of TNF-a, IL-1b, and IL-6 was significantly reduced in Asah1-overexpressing OBA9 cells (39). These results were generally consistent with our findings. The expression of the Asah1 gene is downregulated in neutrophils, and insufficient Asah1 expression will lead to the accumulation of ceramides and the reduction of S1P, resulting in the deterioration and remodeling of the myocardium after MI.
In the final step of de novo SM synthesis, a key gene, Degs1, encodes a desaturase that catalyzes the conversion of dihydroceramide to ceramide by adding a 4,5-trans-double bond (40). The levels of individual dihydroceramide and ceramide species as well as the ratios between species are associated with cardiovascular and metabolic diseases. Elevated ceramide level has been repeatedly shown to be associated with cardiovascular diseases. Gene expression analysis of Degs1-silenced cells revealed dramatic changes in cellular functions, including cell replication, intercellular adhesion, and autophagy (41). A previous study showed that the expression level of Degs1 was significantly increased in granulocyte-monocyte progenitors (GMPs) after 6 h of cytokine stimulation. Furthermore, it has been shown that subsequent activation of Degs1 due to a sudden increase in de novo ceramide production may contribute to reperfusion cardiac injury (42). SPT is considered to be a heterodimer composed of 2 subunits, namely Sptlc1 and Sptlc2. It is the rate-limiting enzyme for sphingolipid biosynthesis. A previous study showed that inhibition of SPT reduced ventricular remodeling, fibrosis, and macrophage content after MI, and deletion of the Sptlc2 gene protected cardiac function after MI. In addition, SPT can also be activated by inflammatory responses by upregulating the expression of Sptlc2 (43). It has been found that Sptlc2 mediates antigenic stimulation and inflammatory signaling, modulates the SM of T cells, maintains the metabolic adaptation of CD8+ T cells, and supports protective immunity (44). Furthermore, neutrophil elastase may increase airway ceramide levels by increasing Sptlc2 protein levels in mouse lungs (32).
Therefore, the pathogenesis of MI involves immune-metabolic disorders. These mechanisms include excessive accumulation of toxic or bioactive lipids as well as disruption of specific key cellular and physiological processes. The SM-related genes Asah1, Degs1, and Sptlc2 are associated with neutrophils and may be potential therapeutic targets for MI. However, these results were obtained from a database-based analysis, and further studies are needed to determine their therapeutic potential in MI. The specific mechanisms by which these genes affect the inflammatory response in MI remain to be further validated experimentally. Our findings may contribute to a deeper understanding of the roles of metabolic genes in the pathogenesis of MI. Reprogramming SM to affect immune cells may become a new modality for MI protective therapy.
In summary, the discovery that the SM genes Asah1, Degs1, and Sptlc2 are highly correlated with immune cell metabolism may deepen our understanding of metabolic genes in the pathogenesis of MI. Reprogramming SM to modulate immune cells may provide a new mode of protective therapy for MI. However, our results were derived from gene expression profiles. Further research and corresponding experimental verification are needed to clarify the specific mechanisms.
Acknowledgments
Funding: This work was funded by the Scientific Research Project of Guangdong Provincial Administration of Traditional Chinese Medicine (No. 20201158).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1041/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1041/coif). WW is from Guangzhou Xidai Hemodialysis Center Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thygesen K, Alpert JS, White HD, et al. Universal definition of myocardial infarction. Circulation 2007;116:2634-53. [Crossref] [PubMed]
- Anderson JL, Morrow DA. Acute Myocardial Infarction. N Engl J Med 2017;376:2053-64. [Crossref] [PubMed]
- Virani SS, Alonso A, Benjamin EJ, et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation 2020;141:e139-596. [Crossref] [PubMed]
- Heusch G, Libby P, Gersh B, et al. Cardiovascular remodelling in coronary artery disease and heart failure. Lancet 2014;383:1933-43. [Crossref] [PubMed]
- Ong SB, Hernández-Reséndiz S, Crespo-Avilan GE, et al. Inflammation following acute myocardial infarction: Multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol Ther 2018;186:73-87. [Crossref] [PubMed]
- Szummer K, Wallentin L, Lindhagen L, et al. Improved outcomes in patients with ST-elevation myocardial infarction during the last 20 years are related to implementation of evidence-based treatments: experiences from the SWEDEHEART registry 1995-2014. Eur Heart J 2017;38:3056-65. [Crossref] [PubMed]
- Andreadou I, Cabrera-Fuentes HA, Devaux Y, et al. Immune cells as targets for cardioprotection: new players and novel therapeutic opportunities. Cardiovasc Res 2019;115:1117-30. [Crossref] [PubMed]
- Veiga-Fernandes H, Freitas AA. The S(c)ensory Immune System Theory. Trends Immunol 2017;38:777-88. [Crossref] [PubMed]
- Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol 2017;17:21-9. [Crossref] [PubMed]
- Abnave P, Ghigo E. Role of the immune system in regeneration and its dynamic interplay with adult stem cells. Semin Cell Dev Biol 2019;87:160-8. [Crossref] [PubMed]
- Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature 2014;510:58-67. [Crossref] [PubMed]
- Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol 2018;19:175-91. [Crossref] [PubMed]
- Laaksonen R, Ekroos K, Sysi-Aho M, et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J 2016;37:1967-76. [Crossref] [PubMed]
- Knapp M, Zendzian-Piotrowska M, Kurek K, et al. Myocardial infarction changes sphingolipid metabolism in the uninfarcted ventricular wall of the rat. Lipids 2012;47:847-53. [Crossref] [PubMed]
- Klyachkin YM, Nagareddy PR, Ye S, et al. Pharmacological Elevation of Circulating Bioactive Phosphosphingolipids Enhances Myocardial Recovery After Acute Infarction. Stem Cells Transl Med 2015;4:1333-43. [Crossref] [PubMed]
- Hadas Y, Vincek AS, Youssef E, et al. Altering Sphingolipid Metabolism Attenuates Cell Death and Inflammatory Response After Myocardial Infarction. Circulation 2020;141:916-30. [Crossref] [PubMed]
- Reforgiato MR, Milano G, Fabriàs G, et al. Inhibition of ceramide de novo synthesis as a postischemic strategy to reduce myocardial reperfusion injury. Basic Res Cardiol 2016;111:12. [Crossref] [PubMed]
- Hua T, Bao Q, He X, et al. Lipidomics Revealed Alteration of Sphingolipid Metabolism During the Reparative Phase After Myocardial Infarction Injury. Front Physiol 2021;12:663480. [Crossref] [PubMed]
- Wixon J, Kell D. The Kyoto encyclopedia of genes and genomes--KEGG. Yeast 2000;17:48-55. [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Amulic B, Cazalet C, Hayes GL, et al. Neutrophil function: from mechanisms to disease. Annu Rev Immunol 2012;30:459-89. [Crossref] [PubMed]
- Liew PX, Kubes P. The Neutrophil's Role During Health and Disease. Physiol Rev 2019;99:1223-48. [Crossref] [PubMed]
- Soehnlein O, Steffens S, Hidalgo A, et al. Neutrophils as protagonists and targets in chronic inflammation. Nat Rev Immunol 2017;17:248-61. [Crossref] [PubMed]
- Espaillat MP, Snider AJ, Qiu Z, et al. Loss of acid ceramidase in myeloid cells suppresses intestinal neutrophil recruitment. FASEB J 2018;32:2339-53. [Crossref] [PubMed]
- Seumois G, Fillet M, Gillet L, et al. De novo C16- and C24-ceramide generation contributes to spontaneous neutrophil apoptosis. J Leukoc Biol 2007;81:1477-86. [Crossref] [PubMed]
- Kumar BV, Connors TJ, Farber DL, Human T. Cell Development, Localization, and Function throughout Life. Immunity 2018;48:202-13. [Crossref] [PubMed]
- Bai A, Guo Y. Acid sphingomyelinase mediates human CD4+ T-cell signaling: potential roles in T-cell responses and diseases. Cell Death Dis 2017;8:e2963. [Crossref] [PubMed]
- Haider N, Boscá L, Zandbergen HR, et al. Transition of Macrophages to Fibroblast-Like Cells in Healing Myocardial Infarction. J Am Coll Cardiol 2019;74:3124-35. [Crossref] [PubMed]
- Ganna A, Salihovic S, Sundström J, et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet 2014;10:e1004801. [Crossref] [PubMed]
- Lin WC, Lin CF, Chen CL, et al. Inhibition of neutrophil apoptosis via sphingolipid signaling in acute lung injury. J Pharmacol Exp Ther 2011;339:45-53. [Crossref] [PubMed]
- Karandashova S, Kummarapurugu AB, Zheng S, et al. Neutrophil elastase increases airway ceramide levels via upregulation of serine palmitoyltransferase. Am J Physiol Lung Cell Mol Physiol 2018;314:L206-14. [Crossref] [PubMed]
- Egom EE, Mamas MA, Clark AL. The potential role of sphingolipid-mediated cell signaling in the interaction between hyperglycemia, acute myocardial infarction and heart failure. Expert Opin Ther Targets 2012;16:791-800. [Crossref] [PubMed]
- Lucki NC, Bandyopadhyay S, Wang E, et al. Acid ceramidase (ASAH1) is a global regulator of steroidogenic capacity and adrenocortical gene expression. Mol Endocrinol 2012;26:228-43. [Crossref] [PubMed]
- Duarte C, Akkaoui J, Yamada C, et al. Elusive Roles of the Different Ceramidases in Human Health, Pathophysiology, and Tissue Regeneration. Cells 2020;9:1379. [Crossref] [PubMed]
- Yuan X, Bhat OM, Lohner H, et al. Downregulation of Lysosomal Acid Ceramidase Mediates HMGB1-Induced Migration and Proliferation of Mouse Coronary Arterial Myocytes. Front Cell Dev Biol 2020;8:111. [Crossref] [PubMed]
- de Carvalho LP, Tan SH, Ow GS, et al. Plasma Ceramides as Prognostic Biomarkers and Their Arterial and Myocardial Tissue Correlates in Acute Myocardial Infarction. JACC Basic Transl Sci 2018;3:163-75. [Crossref] [PubMed]
- Ouyang J, Shu Z, Chen S, et al. The role of sphingosine 1-phosphate and its receptors in cardiovascular diseases. J Cell Mol Med 2020;24:10290-301. [Crossref] [PubMed]
- Azuma MM, Balani P, Boisvert H, et al. Endogenous acid ceramidase protects epithelial cells from Porphyromonas gingivalis-induced inflammation in vitro. Biochem Biophys Res Commun 2018;495:2383-9. [Crossref] [PubMed]
- Xie SZ, Garcia-Prat L, Voisin V, et al. Sphingolipid Modulation Activates Proteostasis Programs to Govern Human Hematopoietic Stem Cell Self-Renewal. Cell Stem Cell 2019;25:639-653.e7. [Crossref] [PubMed]
- Blackburn NB, Michael LF, Meikle PJ, et al. Rare DEGS1 variant significantly alters de novo ceramide synthesis pathway. J Lipid Res 2019;60:1630-9. [Crossref] [PubMed]
- Siddique MM, Li Y, Chaurasia B, et al. Dihydroceramides: From Bit Players to Lead Actors. J Biol Chem 2015;290:15371-9. [Crossref] [PubMed]
- Ji R, Akashi H, Drosatos K, et al. Increased de novo ceramide synthesis and accumulation in failing myocardium. JCI Insight 2017;2:e82922. [Crossref]
- Wu J, Ma S, Sandhoff R, et al. Loss of Neurological Disease HSAN-I-Associated Gene SPTLC2 Impairs CD8+ T Cell Responses to Infection by Inhibiting T Cell Metabolic Fitness. Immunity 2019;50:1218-1231.e5. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


