
High expression of glycolysis-related PGM2 gene in relation to poor prognosis and deficient immune cells infiltration in lung adenocarcinoma: a study based on bioinformatics analysis
Introduction
Based on the latest international cancer figures, lung cancer is the utmost universally diagnosed tumor (11.6% of the overall cases) and the major contributor to cancer-related fatalities (18.4% of the overall malignancy deaths) (1). Primary lung malignancy is typically classified into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). Around 83% of all lung cancer cases are NSCLC and lung adenocarcinoma (LUAD) is the most prevalent subtype of NSCLC. Virtually 80% of NSCLC patients are diagnosed at an advanced phase due to the lack of initial detection biomarkers (2-5), and those with advanced lung cancer have a poor prognosis. LUAD exhibits relatively poor prognosis warranting the need for better predictors of clinical outcome and treatment strategies. Consequently, there is an urgent need to distinguish potential biological markers to improve prognosis of LUAD. Multiple factors participate in the development of LUAD including histological, metabolic, and genetic mechanisms; one of the chief features is energetical glucose metabolism. Compared with normally differentiated cells, the metabolism of cancer cells relies on glycolysis instead of mitochondrial oxidative phosphorylation (OXPHOS), consumes glucose at a higher degree, and yields more lactic acid to meet their higher requirement for nutrients (6-8). The alteration of energy metabolism, referred to as aerobic glycolysis or the Warburg effect, is considered a typical hallmark of malignancy (9-11). The Warburg effect is beneficial to tumor progression, proliferation, and metastasis and plays a crucial role in tumor vitality, hence aerobic glycolysis is a critical energy generation pathway for cancer cells, where the molecules involved may convert the potential biomarkers and therapeutic targets for LUAD (12).
Dysregulated expression of glycolysis-related genes (GRGs) is strongly correlated with the progression of various malignancies, and it is being considered as a possible target for the treatment of tumors. Cui et al. acquired 226 differentially expressed central GRG (DE-GRGs) from The Cancer Genome Atlas (TCGA) database, 8 of which were nominated as central DE-GRG using Cox regression analysis to develop a DE-GRG prognostic model, of which risk score is closely associated with multiple clinical features and is an independent hazard factor affecting prognosis. Phosphoglucomutase 2 (PGM2), is one of the central DE-GRG (13). Zhang et al. similarly uncovered that PGM2 expresses abnormally in clear renal cell carcinoma and performs a crucial assessment function as a GRG for the diagnosis and prognostic evaluation of clear renal cell carcinoma (14).
In the present study, we focused on the function of the GRG PGM2 in LUAD. We evaluated the expression of PGM2 in LUAD tissues and its correlation with LUAD stage, pathologic grade, and prognosis. The function of PGM2 in the infiltration of immune cells in LUAD was also reported in this study. It is anticipated that PGM2 may serve as a prognostic biomarker for LUAD. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1043/rc).
Methods
TCGA database analysis
The information of LUAD patients comprising both RNA sequencing data and clinicopathological factors (including pathological stage, gender, TNM stage, smoking status, age, race, and number pack years smoked) was acquired from the TCGA (https://genomecancer.ucsc.edu/) database, which is a widely used data platform for large-scale cancer genome projects.
Oncomine database analysis and the Human Protein Atlas
The Oncomine database (https://www.oncomine.org/resource/login.html) is the most comprehensive cancer-associated gene microarray platform and cohesive data-resource portal in the world. Oncomine allows users to search for the fullest spectrum of cancer mutations, respective clinical data, and gene expression profiles. We made a comparison of the PGM2 gene expression levels in cancer and normal tissues. By merging several omics methods, the Human Protein Atlas (HPA; https://www.proteinatlas.org/) provides data on all human protein transcripts from unique human samples encompassing cells, tissue, and organs. We obtained PGM2 protein immunohistochemistry in LUAD (https://www.proteinatlas.org/ENSG00000169299-PGM2/pathology/lung+cancer) and normal human samples (https://www.proteinatlas.org/ENSG00000169299-PGM2/tissue/lung) from the HPA (images are available from v21.1.proteinatlas.org). When the fold change (FC) was >1.5 and the P value <0.001, the expression levels were considered substantially different in various tissues. The gene rank criterion was determined to be “top 10%”, where the data type was set to “all”.
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database
The STRING platform (https://string-db.org/) has a large amount of data on protein-protein interactions (PPIs). To investigate PGM2-related proteins, we created a PPI network of PGM2 with the aid of the STRING database. The significant criterion was established as a confidence score greater than 0.7.
Survival and statistical analysis
We classified patients into low- and high-PGM2 expression groups based on their median PGM2 gene expression levels. Relevant gene expression profiles and relevant clinical information was collected by Xiantao platform (www.xiantao.love).With the use of the Kaplan-Meier plotter database (https://kmplot.com/analysis/), we evaluated the possible correlation between PGM2 expression and progression-free interval (PFI), disease-specific survival (DSS), and overall survival (OS) in LUAD patients. The following were the selection thresholds: log-rank P value <0.05 (two-sided) and hazard ratio (HR) within the 95% confidence interval (CI).
Correlation analysis regarding immune cell infiltration
The Tumor Immune Estimation Resource (TIMER) (cistrome.org/TIMER/), a publicly accessible web platform containing data on 32 kinds of cancer and incorporating 10,897 samples from the TCGA database, was utilized to assess the correlation between PGM2 expression and the abundance of 6 infiltrating immune cells (neutrophils, CD4+ T cells, dendritic cells, B cells, macrophages, and CD8+ T cells) among patients with LUAD. We also observed the correlation between PGM2 gene expression and the marker genes of tumor-infiltrating immune cells (TIICs) via the TIMER website.
Quantitative real-time polymerase chain reaction
A total of 12 pairs of clinical LUAD samples and adjacent lung tissues of primary LUAD patients from Wuxi People’s Hospital were collected during surgery and instantly frozen in liquid nitrogen and preserved at −80 °C for further use. Total RNA from tissues was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). The RNA was then reverse-transcribed into complementary DNA (cDNA) according to the manufacturer’s instructions. Then, we performed quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR Green Real-time PCR Master Mix (Thermo Fisher, USA). Relative gene expression was calculated using the 2−DDCq method. The primer sequences were displayed as follows: PGM2, F: GAGGCAGTGAAACGACTAATAGC, R: CTGTCCCAAACTCCATTCGGG; GAPDH, F: CCTTCCGTGTCCCCACT, R: GCCTGCTTCACCACCTTC. This study was approved by the Institutional Research Ethics Committees of Wuxi People’s Hospital Affiliated to Nanjing Medical University (No. KY21075), and informed consent was taken from all the patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The software SPSS (IBM Corp., Armonk, NY, USA) and GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA) were utilized to perform analyses of all statistical data. All data were expressed as means ± standard deviations. For the purpose of determining the significance level of difference across the 2 groups, 2-tailed Student’s t-tests were carried out. Results with P values <0.05 (two-sided) were interpreted as statistically significant unless otherwise specified.
Results
Baseline characteristics of patients
In total, 535 LUAD and 59 normal tissue samples were included in the present study from the TCGA database. Among the 535 participants, 286 patients were female (53.5%) and 249 patients were male (46.5%). Some 446 patients were smokers (85.6%). In regards to the pathological stage, 294 partcipants (55.8%) were already in stage I, 123 patients (23.3%) were in stage II, 84 patients (15.9%) were in stage III, and 26 patients were in stage IV (4.9%). The detailed clinical information is listed in Table 1.
Table 1
| Characteristics | Low expression of PGM2 | High expression of PGM2 | Overall |
|---|---|---|---|
| n | 267 | 268 | 535 |
| T stage, n (%) | |||
| T1 | 105 (19.7) | 70 (13.2) | 175 (32.9) |
| T2 | 127 (23.9) | 162 (30.5) | 289 (54.3) |
| T3 | 25 (4.7) | 24 (4.5) | 49 (9.2) |
| T4 | 9 (1.7) | 10 (1.9) | 19 (3.6) |
| N stage, n (%) | |||
| N0 | 192 (37) | 156 (30.1) | 348 (67.1) |
| N1 | 39 (7.5) | 56 (10.8) | 95 (18.3) |
| N2 | 27 (5.2) | 47 (9.1) | 74 (14.3) |
| N3 | 1 (0.2) | 1 (0.2) | 2 (0.4) |
| M stage, n (%) | |||
| M0 | 178 (46.1) | 183 (47.4) | 361 (93.5) |
| M1 | 9 (2.3) | 16 (4.1) | 25 (6.5) |
| Pathologic stage, n (%) | |||
| Stage I | 165 (31.3) | 129 (24.5) | 294 (55.8) |
| Stage II | 54 (10.2) | 69 (13.1) | 123 (23.3) |
| Stage III | 35 (6.6) | 49 (9.3) | 84 (15.9) |
| Stage IV | 9 (1.7) | 17 (3.2) | 26 (4.9) |
| Primary therapy outcome, n (%) | |||
| PD | 25 (5.6) | 46 (10.3) | 71 (15.9) |
| SD | 21 (4.7) | 16 (3.6) | 37 (8.3) |
| PR | 2 (0.4) | 4 (0.9) | 6 (1.3) |
| CR | 181 (40.6) | 151 (33.9) | 332 (74.4) |
| Gender, n (%) | |||
| Female | 152 (28.4) | 134 (25) | 286 (53.5) |
| Male | 115 (21.5) | 134 (25) | 249 (46.5) |
| Race, n (%) | |||
| Asian | 4 (0.9) | 3 (0.6) | 7 (1.5) |
| Black or African American | 36 (7.7) | 19 (4.1) | 55 (11.8) |
| White | 202 (43.2) | 204 (43.6) | 406 (86.8) |
| Age, n (%) | |||
| ≤65 | 129 (25) | 126 (24.4) | 255 (49.4) |
| >65 | 131 (25.4) | 130 (25.2) | 261 (50.6) |
| No. of pack years smoked, n (%) | |||
| <40 | 99 (26.8) | 89 (24.1) | 188 (50.9) |
| ≥40 | 86 (23.3) | 95 (25.7) | 181 (49.1) |
| Smoker, n (%) | |||
| No | 38 (7.3) | 37 (7.1) | 75 (14.4) |
| Yes | 225 (43.2) | 221 (42.4) | 446 (85.6) |
PGM2, phosphoglucomutase 2.
PGM2 expression level in LUAD and other malignancies
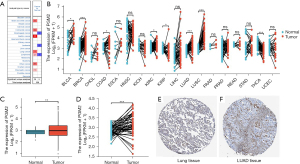
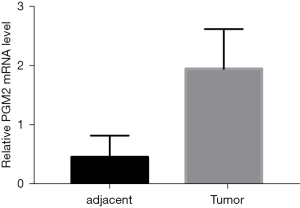
To investigate the involvement of PGM2 in oncogenesis, we performed Oncomine analysis to explore the PGM2 messenger RNA (mRNA) level in different kinds of cancer. The results illustrated that the PGM2 gene was down-regulated in esophageal cancer and leukemia. However, PGM2 gene expression was found to be remarkably upmodulated in lung cancer, central nervous system cancer, pancreatic cancer, brain cancer, gastric cancer, and colorectal cancer. Subsequently, the expression level of PGM2 mRNA in lymphoma was elevated in 2 studies and decreased in 1 study (Figure 1A). Meanwhile, PGM2 was expressed at a high level in 5 kinds of cancers, such as lung squamous cell carcinoma (LUSC) and LUAD based on RNA sequence data found in the TCGA (Figure 1B). The mRNA expression level of PGM2 was significantly elevated in tumor samples as opposed to unpaired normal LUAD samples in TCGA (Figure 1C, P=0.003). The results were verified in LUAD samples and paired normal lung samples (Figure 1D, P=0.001). Moreover, the expression of PGM2 protein was up-regulated in LUAD samples in contrast with normal samples in the HPA (Figure 1E,1F). Besides, we performed qRT-PCR to validate the PGM2 mRNA expression in LUAD and found that PGM2 mRNA was up-regulated in the LUAD tumors (n=12) compared with normal lung tissues (n=12; P<0.001; Figure 2). Overall, these results strongly suggested that PGM2 was up-regulated in LUAD and might perform a negative role in LUAD.
Association of PGM2 expression with clinical features
We assessed the correlation of PGM2 expression with clinical features among patients with LUAD. Elevated expression level of PGM2 was remarkably correlated with T stage (T2 vs. T1, P.adj=0.002) and primary therapeutic outcome (CR vs. PD, P.adj=0.042). There was no remarkable correlation detected between elevated expression level of of PGM2 and other clinical features, including pathological stage, gender, M stage, smoker, age, N stage, race, and number pack years smoked (Figure 3). In univariate logistic regression analysis, elevated expression level of of PGM2 exhibited a substantial correlation with T stage [T2, T3, and T4 vs. T1, odds ratio (OR) =1.826, 95% confidence interval (CI): 1.267–2.644, P=0.001], N stage (N1, N2, and N3 vs. N0, OR =1.910, 95% CI: 1.319–2.782, P<0.001), pathological stage (stage III and stage IV vs. stage I and stage II, OR =1.659, 95% CI: 1.086–2.555, P=0.020), and primary therapeutic outcome [stable disease (SD), partial response (PR), and complete response (CR) vs. progressive disease (PD); OR =0.456, 95% CI: 0.266–0.766, P=0.003), as shown in Table 2.
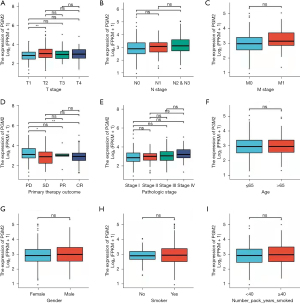
Table 2
| Characteristics | Total (N) | Odds ratio (95% CI) | P value |
|---|---|---|---|
| T stage (T2 & T3 & T4 vs. T1) | 532 | 1.826 (1.267–2.644) | 0.001 |
| N stage (N1 & N2 & N3 vs. N0) | 519 | 1.910 (1.319–2.782) | <0.001 |
| M stage (M1 vs. M0) | 386 | 1.729 (0.759–4.181) | 0.203 |
| Pathologic stage (stage III & stage IV vs. stage I & stage II) | 527 | 1.659 (1.086–2.555) | 0.020 |
| Primary therapy outcome (SD, PR, CR vs. PD) | 446 | 0.456 (0.266–0.766) | 0.003 |
| Gender (male vs. female) | 535 | 1.322 (0.941–1.860) | 0.108 |
| Race (Black or African American & White vs. Asian) | 468 | 1.249 (0.273–6.401) | 0.772 |
| Age (>65 vs. ≤65 years) | 516 | 1.016 (0.719–1.435) | 0.928 |
| Residual tumor (R1 & R2 vs. R0) | 372 | 2.671 (0.969–8.540) | 0.070 |
| Anatomic neoplasm subdivision (right vs. left) | 520 | 1.009 (0.710–1.435) | 0.959 |
| Anatomic neoplasm subdivision 2 (peripheral lung vs. central lung) | 189 | 1.359 (0.739–2.517) | 0.325 |
| Number of pack years smoked (≥40 vs. <40) | 369 | 1.229 (0.817–1.851) | 0.323 |
| Smoker (yes vs. no) | 521 | 1.009 (0.618–1.649) | 0.972 |
PGM2, phosphoglucomutase 2; CI, confidence interval; SD, stable disease; PR, partial response; CR, complete response; PD, progressive disease.
Association of enhanced PGM2 expression and unfavorable prognosis in patients with LUAD
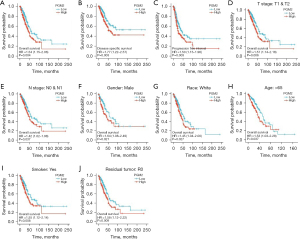
We assessed the prognostic significance of PGM2 in LUAD through Kaplan-Meier analysis. We found that enhanced PGM2 expression was linked to unfavorable patient prognosis including OS (Figure 4A, P=0.004, HR =1.54, 95% CI: 1.15–2.06), DSS (Figure 4B, P=0.003, HR =1.77, 95% CI: 1.22–2.57), and PFI (Figure 4C, P=0.003, HR =1.50, 95% CI: 1.15–1.96) in LUAD. The Kaplan-Meier plotter in LUAD was used to analyze patients belonging to different groups by different clinical variables. The results demonstrated that enhanced PGM2 expression was remarkably linked to shortened OS duration in LUAD cases in the T1/T2 stage (Figure 4D, P=0.006, HR =1.57, 95% CI: 1.14–2.18), N0/N1 satge (Figure 4E, P=0.037, HR =1.42, 95% CI: 1.02–1.98), male (Figure 4F, P=0.021, HR =1.64, 95% CI: 1.08–2.49), white race (Figure 4G, P=0.027, HR =1.45, 95% CI: 1.04–2.00), age over 65 years (Figure 4H, P=0.037, HR =1.53, 95% CI: 1.03–2.28), smokers (Figure 4I, P=0.008, HR =1.55, 95% CI: 1.12–2.14), and with no residual tumor (Figure 4J, P=0.009, HR =1.58, 95% CI: 1.12–2.22). The above findings illustrated the correlation between enhanced PGM2 expression and unfavorable prognosis among patients with LUAD.
Constructing PPIs
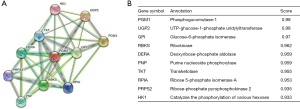
We further sought PPIs to clarify the molecular mechanisms and metabolism of malignant tumors. The PPI network of PGM2 protein was assessed with the help of the STRING tool so as to evaluate their functional interplays in the progression of LUAD. These genes, along with their annotations and scores, are summarized in Figure 5. The 10 most common proteins, including UGP2, GPI, RBKS, DERA, PNP, TKT, RPIA, PRPS2, HK1, and PGM1, participate in both the breakdown and synthesis of glucose. In cellular metabolic pathways, UGP2 serves as a glucose donor.
Assessment of the link between PGM2 expression and immune cells infiltration
Tumor-infiltrating lymphocytes (TILs) have been proposed as a prognostic marker in patients with multiple malignancies. In view of this, the correlation between PGM2 expression of 6 kinds of infiltrating immune cells (neutrophils, macrophages, CD8+ T cells, B cells, dendritic cells, and CD4+ T cells) and tumor purity was analyzed. The findings illustrated a strong correlation between the PGM2 expression and tumor purity (r=−0.096, P=3.29e-02), infiltration levels of B cells (r=−0.154, P=6.71e-04), dendritic cells (r=0.286, P=1.28e-10), CD8+ T cells (r=0.289, P=8.07e-11), neutrophils (r=0.376, P=1.09e-17), macrophages (r=0.286, P=1.34e-10), and CD4+ T cells (r=0.052, P=2.56e-01) in LUAD (Figure 5).
Correlation analysis of PGM2 mRNA expression levels with biomarkers of different immune cell subsets
In addition, we assessed the expression of biomarker genes in diverse immune cells or related subsets in LUAD tissues for the purpose of laying a foundation for future research into PGM2-driven carcinogenesis processes and the identification of viable treatment targets. The findings obtained from TIMER analysis revealed a negative correlation between PGM2 expression and CD19, CD79A, CEACAM8, HLA-DPB1, HLA-DQB1, CD1c, and CCR7. Additionally, a positive correlation was observed between ITGAX, VSIG4, CD163, MS4A4A, IL10, ITGAM, PTGS2, HLA-DRA, IRF5, HLA-DPA1, CD68, NRP1, and CCL2 (Figure 6, Table 3).
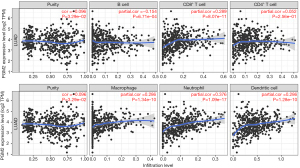
Table 3
| Description | Gene markers | Purity | |
|---|---|---|---|
| Correlation | P value | ||
| B cell | CD19 | −0.168 | 1.75e–04* |
| CD79A | −0.143 | 1.45e–03* | |
| Tumor-associated macrophage | CCL2 | 0.24 | 6.90e–08* |
| CD68 | 0.363 | 8.88e–17* | |
| IL10 | 0.271 | 9.69e–10* | |
| M1 Macrophage | IRF5 | 0.248 | 2.46e–08* |
| PTGS2 | 0.152 | 6.81e–04* | |
| M2 Macrophage | CD163 | 0.361 | 1.16e–16* |
| VSIG4 | 0.275 | 4.94e–10* | |
| MS4A4A | 0.28 | 3.29e–02* | |
| Neutrophils | CEACAM8 | −0.064 | 1.55e–01* |
| ITGAM | 0.25 | 1.86e–08* | |
| CCR7 | −0.033 | 4.61e–01* | |
| Dendritic cell | HLA-DPB1 | −0.016 | 7.19e–01* |
| HLA-DQB1 | −0.085 | 5.98e–02* | |
| HLA-DRA | 0.07 | 1.22e–07* | |
| HLA-DPA1 | 0.048 | 2.91e–01* | |
| CD1c | −0.078 | 8.36e–02* | |
| NRP1 | 0.177 | 7.57e–05* | |
| ITGAX | 0.125 | 5.30e–03* | |
| CD4+ T cells | |||
*P<0.05. PGM2, phosphoglucomutase 2; TIMER, Tumor Immune Estimation Resource.
Discussion
Currently, NSCLC is the most common type of lung cancer which is branded by the dysregulation genetic material such as protein-coding genes and non-coding RNAs (4). Lung cancer is the primary cause of cancer-associated mortality and the survival of this cancer primarily relies upon the phase at diagnosis, though almost two-thirds of lung cancer patients are diagnosed at advanced stages. Our study revealed that the PGM2 gene was remarkably up-regulated in LUAD, which was correlated with differential OS and poor prognosis. Our study similarly recommended that PGM2 is associated with immune implications in LUAD, which may be the mechanism through which PGM2 influences prognosis. The results of our research revealed that PGM2 may become a prospective biomarker for diagnosis and prognosis which may serve an innovative immune-related therapeutic purpose.
The gene PGM2 is one of the central DE-GRG (14). The results in our study suggested that PGM2 was up-regulated in LUAD and might play a negative role in LUAD, which indicates that the expression of PGM2 can distinguish LUAD patients from healthy individuals with extraordinary sensitivity and specificity. The elevated expression level of PGM2 was remarkably linked to T stage, especially in T1, T2, and primary therapeutic outcome, but there was no remarkable correlation between the elevated expression level of PGM2 and other clinical features, including M stage, pathologic stage, age, gender, smoking history, race, and number of pack-years smoked. These results indicate that PGM2 has a vast diagnostic value for LUAD at the initial phase.
Kaplan-Meier analysis was performed for the purpose of assessing PGM2’s prognostic significance in LUAD. Moreover, we discovered that enhanced PGM2 expression was linked to an unfavorable prognosis in patients with LUAD, as validated by the values of DSS, OS, and PFI. The findings from subgroup analysis illustrated that enhanced expression of PGM2 was substantially related to an unfavorable OS in LUAD cases in the T1/T2 stage, N0/N1 satge, and so on. Multivariate and univariate analyses revealed that enhanced expression of PGM2 may be described as a hazard element that influences the OS of patients with LUAD. Thus, we confirmed that PGM2 has prognostic value in LUAD.
Recently, with the upsurge of immunotherapy, the growth of immune-checkpoint inhibitors (ICIs) has intensely transformed the NSCLC medicinal domain. The progress of immunotherapeutic strategies transforms the stability of immune viability away from a cancer-induced immune oppressive state to a vigorous antitumor immunity response (15,16). The findings from the present study illustrated a correlation between the expression of PGM2 and tumor purity, as well as infiltrating levels of macrophages, CD4+ T cells, dendritic cells, B cells, CD8+ T cells, and neutrophils in LUAD. By performing TIMER analysis, we discovered a negative correlation between PGM2 expression and CD19, HLA-DQB1, CEACAM8, CD79A, HLA-DPB1, CD1c, and CCR7. Additionally, a positive correlation was observed between ITGAX, IL10, HLA-DPA1, PTGS2, ITGAM, VSIG4, CD163, MS4A4A, IRF5, HLA-DRA, CD68, NRP1, and CCL2. The T lymphocytes are the most common immune cell types in the LUAD cancer microenvironment, followed by B cells, natural killer cells, and macrophages (17). Dendritic cells are individual antigen-presenting cells that perform a specific function to the incentive of anticancer T lymphocytes (18,19). The influence of T cells universally predicts a superior theoretical consequence in LUAD (20). Additionally, the function of cancer-infiltrating B cells in the microenvironment of cancer has triggered growing responsiveness. The majority of studies point out that the influence of B cells in LUAD is linked to a satisfactory consequence (21,22).
In conclusion, we discovered that PGM2 is upregulated in LUAD tissues and that elevated expression level of PGM2 is correlated with clinical progression and can serve as a risk factor for OS in LUAD patients in an independent manner. Immunotherapy has been the most significant advancement in LUAD treatment. The findings of the present study illustrated that a higher expression level of the glycolysis-related PGM2 gene was associated with lower immune cell infiltration. Moreover, PGM2 might influence glycolysis pathways in LUAD, which might be crucial in cancer. Additionally, PGM2 might serve as a marker for helping to identify patients who may be candidates for glycolysis-induction therapy or its integration with immunotherapy. Taken together, PGM2 is a potential biological marker for predicting the prognosis of patients with LUAD.
Acknowledgments
Funding: This work was supported by the Wuxi Health Committee (No. T202034).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1043/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1043/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1043/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Research Ethics Committees of Wuxi People’s Hospital Affiliated to Nanjing Medical University (No. KY21075), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Aggarwal C, Rolfo CD, Oxnard GR, et al. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat Rev Clin Oncol 2021;18:56-62. [Crossref] [PubMed]
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol 2021;157:103194. [Crossref] [PubMed]
- Teng PC, Liang Y, Yarmishyn AA, et al. RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases. Int J Mol Sci 2021;22:10592. [Crossref] [PubMed]
- Passaro A, Leighl N, Blackhall F, et al. ESMO expert consensus statements on the management of EGFR mutant non-small-cell lung cancer. Ann Oncol 2022;33:466-87. [Crossref] [PubMed]
- Jiang H, Wei H, Wang H, et al. Zeb1-induced metabolic reprogramming of glycolysis is essential for macrophage polarization in breast cancer. Cell Death Dis 2022;13:206. [Crossref] [PubMed]
- Liu Y, Tang W, Ren L, et al. Activation of miR-500a-3p/CDK6 axis suppresses aerobic glycolysis and colorectal cancer progression. J Transl Med 2022;20:106. [Crossref] [PubMed]
- Guo JQ, Wang CD, Tang HY, et al. PDGF-BB/PDGFRβ promotes epithelial-mesenchymal transition by affecting PI3K/AKT/mTOR-driven aerobic glycolysis in Wilms' tumor G401 cells. Cell Biol Int 2022;46:907-21. [Crossref] [PubMed]
- Chen Y, Song S, Zhang L, et al. Circular RNA hsa_circ_0091579 facilitates the Warburg effect and malignancy of hepatocellular carcinoma cells via the miR-624/H3F3B axis. Clin Transl Oncol 2021;23:2280-92. [Crossref] [PubMed]
- Krisher RL, Prather RS. A role for the Warburg effect in preimplantation embryo development: metabolic modification to support rapid cell proliferation. Mol Reprod Dev 2012;79:311-20. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, et al. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol 2017;14:113. [Crossref] [PubMed]
- Cui Z, Sun G, Bhandari R, et al. Comprehensive Analysis of Glycolysis-Related Genes for Prognosis, Immune Features, and Candidate Drug Development in Colon Cancer. Front Cell Dev Biol 2021;9:684322. [Crossref] [PubMed]
- Zhang Y, Chen M, Liu M, et al. Glycolysis-Related Genes Serve as Potential Prognostic Biomarkers in Clear Cell Renal Cell Carcinoma. Oxid Med Cell Longev 2021;2021:6699808. [Crossref] [PubMed]
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. [Crossref] [PubMed]
- Carbone DP, Gandara DR, Antonia SJ, et al. Non-Small-Cell Lung Cancer: Role of the Immune System and Potential for Immunotherapy. J Thorac Oncol 2015;10:974-84. [Crossref] [PubMed]
- Stankovic B, Bjørhovde HAK, Skarshaug R, et al. Immune Cell Composition in Human Non-small Cell Lung Cancer. Front Immunol 2018;9:3101. [Crossref] [PubMed]
- Wang JB, Huang X, Li FR. Impaired dendritic cell functions in lung cancer: a review of recent advances and future perspectives. Cancer Commun (Lond) 2019;39:43. [Crossref] [PubMed]
- Wculek SK, Cueto FJ, Mujal AM, et al. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol 2020;20:7-24. [Crossref] [PubMed]
- Zhang J, Endres S, Kobold S. Enhancing tumor T cell infiltration to enable cancer immunotherapy. Immunotherapy 2019;11:201-13. [Crossref] [PubMed]
- Diao XY, Wang F. Single-cell transcriptome analysis of tumor-infiltrating B cells reveals their clinical implications in non-small cell lung cancer. Thorac Cancer 2021;12:5-7. [Crossref] [PubMed]
- Wu J, Li L, Zhang H, et al. A risk model developed based on tumor microenvironment predicts overall survival and associates with tumor immunity of patients with lung adenocarcinoma. Oncogene 2021;40:4413-24. [Crossref] [PubMed]
(English Language Editor: J. Jones)


