
Structure-based classification of EGFR mutations in operable pre-invasive and invasive non-small cell lung cancer: a cross-sectional study
Introduction
Lung cancer is the most common cause of cancer death. It is broadly divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), with 80–85% classified as NSCLC (1). Although up to 30% of patients with NSCLC can be diagnosed early and undergo curative surgery, disease recurrence is still common in early-stage disease (2,3). Nearly half of patients with stage IB NSCLC and more than three-quarters of patients with stage IIIA NSCLC experience recurrence within 5 years (4). Adjuvant treatment is recommended for patients to reduce the risk of postoperative recurrence (5-7).
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are a targeted first-line treatment for patients with EGFR mutation–positive advanced NSCLC (8-11). Several studies have shown that EGFR-TKIs as adjuvant therapy improve the prognosis of early-stage patients with EGFR exon 19 deletions and L858R mutations (12-14). In addition to classical EGFR mutations, atypical EGFR mutations have been identified in 10–30% of patients with NSCLC (15-17). However, the use of EGFR-TKI treatment for patients with atypical EGFR mutations has not been well-studied. To understand the effect of atypical EGFR mutations on patient outcome, a recent study proposed a structure-based approach for improving the prediction of drug sensitivity in patients with atypical EGFR mutations (18). Four EGFR mutation subgroups were identified based on structure–function using a drug sensitivity assay and an in silico prediction model: (I) classical-like mutations that were distant from the ATP-binding pocket and were predicted to have little effect on the overall structure of EGFR, including L861Q, T725M, and EGFR classical mutations. These mutations were sensitive for all classes of EGFR-TKIs; (II) exon 20 insertions (Ex20ins), insertions in the loop at the C-terminal end of the α C-helix in exon 20, which can be subdivided into near-loop (NL) and far-loop (FL) insertions based on in vitro sensitivity. Second-generation TKIs and Ex20ins-active TKIs were more sensitive in Ex20ins-NL than in Ex20ins-FL; (III) mutations on the interior surface of the ATP-binding pocket or C-terminal end of the α C-helix, which were predicted to be P-loop and α C-helix compressing (PACC), including G719A and E709A. PACC mutations were more sensitive to second-generation TKIs than any other TKI class; (IV) T790M-like mutations in the hydrophobic core, which were mostly composed of complex mutations combined with T790M mutations. T790M-like mutations consist of 2 subgroups of third-generation TKI-sensitive (T790M-like-3S) and third-generation TKI-resistant (T790M-like-3R) mutations.
Adenocarcinoma in situ (AIS) is a subtype of NSCLC, which exhibits early-stage growth patterns but can develop into invasion (19). The 10-year recurrence-free survival of AIS is 100% with appropriate therapy, and the 10-year overall survival is 98.1% (20). However, the 5‐year overall survival rate of advanced NSCLC patients is less than 7% (21). While there was a great amount of data about EGFR mutations in advanced NSCLC, less is known about AIS. In the study of genomic and immune profiling of pre-invasive lung adenocarcinoma, 28 AIS patients were included (22). Another study analyzed the mutational profile of Chinese NSCLC patients of adenocarcinoma including 21 AIS patients (23). Nakamura et al. (24) reported the EGFR mutation rates in earliest phases of lung adenocarcinoma with 22 AIS and 21 minimally invasive adenocarcinoma (MIA). In this study, we were able to include 66 AIS patients as a subgroup of pre-invasive NSCLC for the structural analysis of EGFR mutations.
In total, we analyzed 1,012 NSCLC patients with pathological stage I–III or AIS to evaluate relationship of the clinical characteristics and EGFR mutations stratified either by traditional exon-based method or the structure-based approach. We present the following article in accordance with the STROBE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1054/rc).
Methods
Patients and tissue samples
To systemically analyze the structure-based classification of EGFR mutations in operable NSCLC patients, we performed this retrospective descriptive cross-section study. After excluding patients with stage IV or without EGFR mutations at stage I–III were excluded, 1,012 stage I–III NSCLC patients with EGFR mutation tested with next-generation sequencing (NGS) from Tongxiang First People’s Hospital, First Medical Center of PLA General Hospital and Affiliated Qingdao Central Hospital between May 2018 and October 2021 were recruited. Clinical data were collected from the medical records of each patient, including gender, age, smoking status, histological subtype, and disease stage at diagnosis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of the Tongxiang First People’s Hospital (No. 2022-002-01) and informed consent was taken from all the patients.
DNA extraction and NGS
Formalin-fixed paraffin-embedded (FFPE) slides were stored at room temperature. Genomic DNA was extracted from FFPE tumor samples using a QIAamp DNA FFPE Tissue Kit according to the manufacturer’s protocol (Qiagen GmbH, Hilden, Germany). DNA from leukocytes was extracted using a DNeasy Blood Kit (Qiagen). DNA concentration and size distribution were estimated using a Qubit Fluorometer and a Qubit dsDNA HS Assay Kit according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA).
Sequencing library preparation and sequencing protocol were conducted as described previously (25,26). Briefly, genomic DNA libraries were constructed with a KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA). The capture probe design was based on approximately 1.45 Mb genomic regions of 1,021 genes frequently mutated in solid tumors. DNA sequencing was performed using Gene + Seq-2000 (Geneplus, Beijing, China) with paired-end reads. Matched peripheral blood was sequenced as a control to filter germline variation.
EGFR mutation analysis
Terminal adaptor sequences and low-quality data were removed. The clean reads were aligned to the human genome build GRCh37 using the Burrows-Wheeler Aligner (version 0.7.12-r1039; http://bio-bwa.sourceforge.net/). Single nucleotide variants (SNVs) and insertions or deletions (indels) were identified using GATK (version 3.4-46-gbc02625; Broad Institute, Cambridge, MA, USA) and MuTect (version 1.1.4; Broad Institute). All final candidate variants were verified using an integrative genomics viewer browser. The exon-based EGFR mutation types were categorized into 8 subgroups: Ex19del, L858R, T790M, classical + T790M, Ex20ins, other atypical, complex atypical, and Ex19del + L858R. Using the structure-based approach (18), the following 4 EGFR mutation subgroups were established: classical-like group, PACC group, Ex20ins group, and T790M-like group.
Statistical analysis
Differences among subgroups stratified by stage, gender, age, and smoking status were analyzed by Fisher’s exact test, where appropriate. All analyses were performed using SPSS version 26.0 (IBM Corp., Armonk, NY, USA). Two-sided P values of <0.05 were considered statistically significant.
Results
Clinical characteristics and EGFR mutations
A total of 632 female and 380 male NSCLC patients with a mean age of 61 years (range, 21–96 years) were included in this study. The clinical characteristics of the patients are listed in Table 1. A total of 66 patients (6.52%) with AIS were included in this research. A total of 1,185 EGFR mutations were identified in the 1,012 patients with NSCLC, of whom 84.39% and 15.61% harbored a single EGFR mutation and complex EGFR mutations, respectively. As shown in Figure 1A, L858R and 19del were the major types; as expected, L858R was more common than 19del in our population (39.33% vs. 35.67%). In addition, EGFR T790M mutation was identified in 18 patients (1.78%), including 1 patient with a T790M mutation but no other EGFR mutations. Concurrent L858R and 19del mutations were identified in 9 patients (0.89%), who were all diagnosed with multiple primary lung cancer. The detailed clinical and molecular data of the 9 patients are shown in Table 2.
Table 1
| Clinical factors | Number (%) |
|---|---|
| Gender | |
| Female | 632 (62.45) |
| Male | 380 (37.55) |
| Age (years) | |
| Median [range] | 61 [21–96] |
| <65 | 616 (60.87) |
| ≥65 | 396 (39.13) |
| Stage | |
| 0 | 66 (6.52) |
| I–III | 946 (93.48) |
| Smoking status | |
| Smokers | 132 (13.04) |
| Nonsmokers | 504 (49.80) |
| Unknown | 376 (37.16) |
| Histology | |
| Adenocarcinoma | 916 (90.51) |
| Squamous-cell carcinoma | 17 (1.68) |
| Adenocarcinoma in situ | 66 (6.52) |
| Other types of NSCLC | 13 (1.29) |
NSCLC, non-small cell lung cancer.
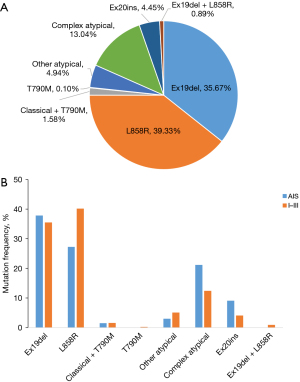
Table 2
| Patient ID | Gender | Age | Stage | EGFR mutation | Tumor site | Other mutations |
|---|---|---|---|---|---|---|
| E0028 | Female | 67 | I | L858R | Left upper lobe | LRP1B p.R1619H; MLL3 p.A4152V; SMO p.A324T |
| E0028 | Female | 67 | I | E746_T751delinsV | Left upper lobe | TMPRSS2 p.I521L |
| E0155 | Female | 65 | Ib | E746_A750del | Right upper lobe | CTNNB1 p.S37F; KRAS p.G12S; RARA p.M284V; SMAD4 p.Q334* |
| E0155 | Female | 65 | Ib | L858R | Left lower lobe | ATRX p.M2492L; RAD51B p.S175I |
| E0183 | Male | 74 | I | L747_T751del | Right upper lobe | ABL1 p.R1095W; ACIN1 p.E274G; MAP3K1 p.V1435G; TP53 p.H193D |
| E0183 | Male | 74 | I | L858R | Right upper lobe | AXL p.A210T; GAB2 p.W661*; HNRPDL p.M120V; PIK3C2B p.D1537_P1538[2>1]; SETD2 p.T904I; SLX4 p.P1095T |
| E0212 | Female | 59 | Ia | L747_T751del | Right lower lung | No |
| E0212 | Female | 59 | Ia | L858R | Right upper lobe | No |
| E0446 | Female | 52 | II | L858R | Right upper lobe | No |
| E0446 | Female | 52 | II | E746_A750del | Right upper lobe | FANCM p.A141S; PTPRD p.L503V |
| E0735 | Female | 66 | I | E746_A750del | Left upper lobe | ARAF p.K336N; ATM p.V2951D; DOT1L p.W611R; DOT1L p.E1360D |
| E0735 | Female | 66 | I | L858R | Left upper lobe | RBM10 p.Q843*; TXNIP p.V54Sfs*20 |
| E0790 | Female | 53 | II | S752_I759del | Left lower lobe | CSF1R p.N240S; RBM10 p.E624* |
| E0790 | Female | 53 | II | L858R | Right upper lobe | RBM10 p.E578*; SETD2 p.Q2334* |
| E0790 | Female | 53 | II | L858R | Left lower lobe | No |
| E0897 | Female | 66 | I | L858R | Right middle lobe | CYP2D6 p.D100E; MED12 p.S440T; NF1 p.D1091V; RBM10 p.E494* |
| E0897 | Female | 66 | I | E746_A750del | Left upper lobe | AXIN1 p.A443V |
| E0966 | Female | 70 | I | E746_A750del | Left upper lobe | WT1 p.Q238R |
| E0966 | Female | 70 | I | L858R | Left upper lobe | EPHA3 p.D316G |
*, translation stop codon. EGFR, epidermal growth factor receptor.
AIS differed from stage I–III NSCLC in terms of exon-based EGFR mutation classification
To comprehensively investigate the EGFR mutation characterization of early-stage NSCLC, 66 patients with AIS and 946 patients with stage I–III NSCLC were included in the study. The mutation profiles of EGFR in the AIS and stage I–III patients are listed in Figure 1B. A different EGFR mutation distribution was observed between AIS and stage I–III patients. As shown in Figure 1B, 37.88%, 27.27%, and 33.33% AIS patients harbored Ex19del, L858R, and atypical mutations, respectively. In the stage I–III group, the distribution of EGFR mutations was 35.52% (Ex19del), 40.17% (L858R), and 21.67% (atypical; P=0.03). The proportion of L858R in AIS patients was lower than that in stage I–III patients, while the proportion of atypical mutations was higher in AIS patients.
Structural-based EGFR mutation classification
Based on a previous publication, we classified EGFR mutations of patients with pre-invasive and invasive NSCLC into 4 distinct subgroups with structural features: (I) classical-like; (II) PACC; (III) Ex20ins, including Ex20ins-NL and Ex20ins-FL; and (IV) T790M-like (Figure 2A). Classical-like mutations were the largest subgroup of EGFR mutations (871 patients, 86.07% of the cohort), followed by PACC (72 patients, 7.11%), Ex20ins (51 patients, 5.04%), and T790M-like (18 patients, 1.78%). The patents of Ex20ins-NL group were more than the Ex20ins-FL group (3.95% vs. 1.09%). The frequency of Ex20ins-NL and Ex20ins-FL mutations is shown in the lollipop plot in Figure 2B. The clinical characteristics and subgroups of EGFR mutations in patients are presented in Table 3. Our analyses indicated that the composition of EGFR mutations was different between patients <65 and ≥65 years (P=0.0267) but similar between patients with AIS and patients with stage I–III NSCLC (P=0.1436). However, a higher percentage of Ex20ins occurred in younger (<65 years) patients, nonsmoking patients, and AIS patients (6.7% vs. 2.5%, P=0.003; 5.8% vs. 0.8%, P=0.0107; and 10.6% vs. 4.7%, P=0.0423 respectively).
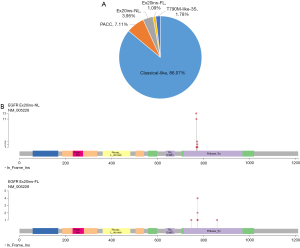
Table 3
| Clinical factors | Classical-like (N=871) | PACC (N=72) | Ex20ins (N=51) | T790M-like (N=18) | P value |
|---|---|---|---|---|---|
| Sex | 0.1325 | ||||
| Female | 550 | 39 | 35 | 8 | |
| Male | 321 | 33 | 16 | 10 | |
| Age (years) | 0.0267 | ||||
| <65 | 522 | 41 | 41 | 12 | |
| ≥65 | 349 | 31 | 10 | 6 | |
| Stage | 0.1436 | ||||
| AIS | 52 | 5 | 7 | 2 | |
| I–III | 819 | 67 | 44 | 16 | |
| Smoking status | |||||
| Smokers | 115 | 12 | 1 | 4 | – |
| Nonsmokers | 444 | 24 | 29 | 7 | |
| Unknown | 312 | 36 | 21 | 7 | |
| Histology | |||||
| Adenocarcinoma | 794 | 64 | 44 | 14 | – |
| Squamous-cell carcinoma | 12 | 3 | 0 | 2 | |
| AIS | 52 | 5 | 7 | 2 | |
| Other types of NSCLC | 13 | 0 | 0 | 0 |
The associations of EGFR mutation type with clinical variables were evaluated by Fisher’s exact test. P<0.05 was considered statistically significant, and all tests were two-tailed. EGFR, epidermal growth factor receptor; PACC, P-loop and α C-helix compressing; Ex20ins, exon 20 insertions; AIS, adenocarcinoma in situ; NSCLC, non-small cell lung cancer.
Discussion
Research on EGFR-TKI treatment for early-stage NSCLC patients with atypical EGFR mutations is lacking. To our knowledge, this is the first study to delineate the structural classification of EGFR mutations in early-stage NSCLC and AIS using a large cohort.
In our cohort, L858R was more common than 19del, which was similar to previous studies in East Asian patients (27,28) and different to previous study in Western patient populations (18). The atypical mutations were also less common in our cohort than in Western patients (22.43% vs. 30.8%) (18). When classified with traditional exon-based method, patients with AIS exhibited a higher proportion of atypical EGFR mutations than patients with stage I–III lung cancer (33.33% vs. 21.66%; P=0.03). However, according to the structure-based method, the proportion of atypical EGFR mutations in patients with AIS was similar to those with stage I–III lung cancer. In addition, the mutation rate of Ex20ins differed in patients according to age, smoking, and invasion stage. Moreover, we reported 9 patients with Ex19del + L858R double mutations who were diagnosed with synchronous multiple primary lung cancer. As all our patients were EGFR-TKI naïve, there were no T790M-like-3R mutations, which are rare in EGFR-TKI naïve patients but common in 3rd generation EGFR-TKI treated patients (18).
Previous research has indicated that the frequency of EGFR mutation is about 27.3–52% in pre-invasive lung adenocarcinoma, suggesting that EGFR mutations may be an early genetic event in the development of lung cancer (23,24,29,30). Our study included 66 patients with AIS, and the EGFR mutation spectrum was as follows: Ex19del mutation (37.88%), L858R mutation (27.27%), and atypical mutation (33.33%). A retrospective study with 28 AIS patients identified 11 EGFR mutations in 10 samples, including 6 patients with Ex19del, 2 patients with L858R, and 2 patients with atypical mutations (30). Another study identified 4 Ex19del mutations, 4 L858R mutations, 1 Ex19del + L858R double mutation, and 1 other mutation in 10 patients with EGFR mutation-positive atypical adenomatous hyperplasia (AAH)/AIS (31). The different EGFR mutation rate between our patients and previous studies may have been caused by limited sample size or the method of EGFR detection.
Patients harboring EGFR Ex20ins exhibited poorer prognosis compared to patients with sensitizing mutations in EGFR. Ex20ins mutations are heterogenous in EGFR-TKI (32,33). Previous studies have demonstrated that patients with EGFR Ex20ins mutations are usually non-smoking females (34-37). Arcila et al. reported that among 33 patients with EGFR Ex20ins, 67% were female, 48% had never smoked, 55% were in stage I–II, and 45% were in stage III–IV (34). EGFR Ex20ins were more common among non-smoking patients (P<0.0001), and no significant difference was detected in age, sex, race, or stage. Another study with 27 patients with EGFR Ex20ins found that 19 patients were females (P=0.24), 15 patients had never smoked (P<0.001), 8 patients were in stage I–III, and 19 patients were in stage IV (P=0.05) (35). Our study further demonstrated that in patients with early-stage NSCLC, Ex20ins occur in patients who are younger, nonsmoking, and have AIS (P=0.003, P=0.017, and P=0.0423, respectively).
There were several limitations in our study. Firstly, it was a retrospective analysis, and thus no EGFR-TKI adjuvant therapy data were included. Secondly, only AIS data were available in our cohort, and thus AAH and MIA data were not included. Finally, the smoking status of some patients was not clear.
In summary, we have delineated the structural classification of EGFR mutations in early lung cancer and AIS using a large cohort. Whether this approach can improve predictions of targeted therapy efficacy in adjuvant therapy is worthy of further study.
Acknowledgments
The authors wish to acknowledge the study staff, the study patients, and their families.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1054/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1054/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1054/coif). LW, YX, and RC are employees of Geneplus-Beijing Institute. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of the Tongxiang First People’s Hospital (No. 2022-002-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Xie S, Wu Z, Qi Y, et al. The metastasizing mechanisms of lung cancer: Recent advances and therapeutic challenges. Biomed Pharmacother 2021;138:111450. [Crossref] [PubMed]
- Chmielewska I, Stencel K, Kalinka E, et al. Neoadjuvant and Adjuvant Immunotherapy in Non-Small Cell Lung Cancer-Clinical Trials Experience. Cancers (Basel) 2021;13:5048. [Crossref] [PubMed]
- Galvez C, Jacob S, Finkelman BS, et al. The role of EGFR mutations in predicting recurrence in early and locally advanced lung adenocarcinoma following definitive therapy. Oncotarget 2020;11:1953-60. [Crossref] [PubMed]
- Mithoowani H, Febbraro M. Non-Small-Cell Lung Cancer in 2022: A Review for General Practitioners in Oncology. Curr Oncol 2022;29:1828-39. [Crossref] [PubMed]
- Bradbury P, Sivajohanathan D, Chan A, et al. Postoperative Adjuvant Systemic Therapy in Completely Resected Non-Small-Cell Lung Cancer: A Systematic Review. Clin Lung Cancer 2017;18:259-273.e8. [Crossref] [PubMed]
- Lim JU, Yeo CD. Update on adjuvant therapy in completely resected NSCLC patients. Thorac Cancer 2022;13:277-83. [Crossref] [PubMed]
- Passiglia F, Bertaglia V, Reale ML, et al. Major breakthroughs in lung cancer adjuvant treatment: Looking beyond the horizon. Cancer Treat Rev 2021;101:102308. [Crossref] [PubMed]
- Hanna N, Johnson D, Temin S, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2017;35:3484-515. [Crossref] [PubMed]
- Khaddour K, Jonna S, Deneka A, et al. Targeting the Epidermal Growth Factor Receptor in EGFR-Mutated Lung Cancer: Current and Emerging Therapies. Cancers (Basel) 2021;13:3164. [Crossref] [PubMed]
- Moore S, Wheatley-Price P. EGFR Combination Therapy Should Become the New Standard First-Line Treatment in Advanced EGFR-Mutant NSCLC. J Thorac Oncol 2021;16:1788-92. [Crossref] [PubMed]
- Hayashi H, Nadal E, Gray JE, et al. Overall Treatment Strategy for Patients With Metastatic NSCLC With Activating EGFR Mutations. Clin Lung Cancer 2022;23:e69-82. [Crossref] [PubMed]
- Wu YL, Tsuboi M, He J, et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:1711-23. [Crossref] [PubMed]
- Lin C, Hu F, Chu H, et al. The role of EGFR-TKIs as adjuvant therapy in EGFR mutation-positive early-stage NSCLC: A meta-analysis. Thorac Cancer 2021;12:1084-95. [Crossref] [PubMed]
- Frampton JE. Osimertinib: A Review in Completely Resected, Early-Stage, EGFR Mutation-Positive NSCLC. Target Oncol 2022;17:369-76. [Crossref] [PubMed]
- Yang JC, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Passaro A, Mok T, Peters S, et al. Recent Advances on the Role of EGFR Tyrosine Kinase Inhibitors in the Management of NSCLC With Uncommon, Non Exon 20 Insertions, EGFR Mutations. J Thorac Oncol 2021;16:764-73. [Crossref] [PubMed]
- Harrison PT, Vyse S, Huang PH. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol 2020;61:167-79. [Crossref] [PubMed]
- Robichaux JP, Le X, Vijayan RSK, et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021;597:732-7. [Crossref] [PubMed]
- Li D, Yang W, Zhang Y, et al. Genomic analyses based on pulmonary adenocarcinoma in situ reveal early lung cancer signature. BMC Med Genomics 2018;11:106. [Crossref] [PubMed]
- Li D, Deng C, Wang S, et al. Ten-year follow-up of lung cancer patients with resected adenocarcinoma in situ or minimally invasive adenocarcinoma: Wedge resection is curative. J Thorac Cardiovasc Surg 2022;S0022-5223(22)00713-9. Epub ahead of print. [Crossref] [PubMed]
- Li F, Dong X. Pembrolizumab provides long-term survival benefits in advanced non-small cell lung cancer: The 5-year outcomes of the KEYNOTE-024 trial. Thorac Cancer 2021;12:3085-7. [Crossref] [PubMed]
- Chen H, Carrot-Zhang J, Zhao Y, et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun 2019;10:5472. [Crossref] [PubMed]
- Ding Y, Zhang L, Guo L, et al. Comparative study on the mutational profile of adenocarcinoma and squamous cell carcinoma predominant histologic subtypes in Chinese non-small cell lung cancer patients. Thorac Cancer 2020;11:103-12. [Crossref] [PubMed]
- Nakamura H, Koizumi H, Kimura H, et al. Epidermal growth factor receptor mutations in adenocarcinoma in situ and minimally invasive adenocarcinoma detected using mutation-specific monoclonal antibodies. Lung Cancer 2016;99:143-7. [Crossref] [PubMed]
- Ai X, Cui J, Zhang J, et al. Clonal Architecture of EGFR Mutation Predicts the Efficacy of EGFR-Tyrosine Kinase Inhibitors in Advanced NSCLC: A Prospective Multicenter Study (NCT03059641). Clin Cancer Res 2021;27:704-12. [Crossref] [PubMed]
- Xu J, Wang R, Wang T, et al. Targeted DNA profiling and the prevalence of NTRK aberrations in Chinese patients with head and neck cancer. Oral Oncol 2021;119:105369. [Crossref] [PubMed]
- Yotsukura M, Yasuda H, Shigenobu T, et al. Clinical and pathological characteristics of EGFR mutation in operable early-stage lung adenocarcinoma. Lung Cancer 2017;109:45-51. [Crossref] [PubMed]
- Pi C, Xu CR, Zhang MF, et al. EGFR mutations in early-stage and advanced-stage lung adenocarcinoma: Analysis based on large-scale data from China. Thorac Cancer 2018;9:814-9. [Crossref] [PubMed]
- Zhang C, Zhang J, Xu FP, et al. Genomic Landscape and Immune Microenvironment Features of Preinvasive and Early Invasive Lung Adenocarcinoma. J Thorac Oncol 2019;14:1912-23. [Crossref] [PubMed]
- Xu X, Li N, Zhao R, et al. Targeted next-generation sequencing for analyzing the genetic alterations in atypical adenomatous hyperplasia and adenocarcinoma in situ. J Cancer Res Clin Oncol 2017;143:2447-53. [Crossref] [PubMed]
- Liu M, He WX, Song N, et al. Discrepancy of epidermal growth factor receptor mutation in lung adenocarcinoma presenting as multiple ground-glass opacities. Eur J Cardiothorac Surg 2016;50:909-13. [Crossref] [PubMed]
- Kosaka T, Tanizaki J, Paranal RM, et al. Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res 2017;77:2712-21. [Crossref] [PubMed]
- Robichaux JP, Elamin YY, Tan Z, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 2018;24:638-46. [Crossref] [PubMed]
- Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 2013;12:220-9. [Crossref] [PubMed]
- Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013;8:179-84. [Crossref] [PubMed]
- Byeon S, Kim Y, Lim SW, et al. Clinical Outcomes of EGFR Exon 20 Insertion Mutations in Advanced Non-small Cell Lung Cancer in Korea. Cancer Res Treat 2019;51:623-31. [Crossref] [PubMed]
- Yang S, Jiang Z, Lu H. Clinical and pathological findings of surgically resected patients for lung adenocarcinomas harboring uncommon EGFR mutations. Int J Clin Exp Pathol 2017;10:7627-31. [PubMed]
(English Language Editor: C. Gourlay)


