
Accurate detection of lung cancer-related microRNA through CRISPR/Cas9-assisted garland rolling circle amplification
Introduction
Lung cancer remains the most threatening cause of fatal malignancies, with approximately 1.6 million deaths per annum globally. Clinical success of therapies is poor due to delayed diagnosis, restricted therapeutic tools, relapse, and drug resistance. The main cause of death in lung cancer is metastasis, and the brain is one of the most common metastatic sites of lung cancer. Research shows that more than 50% of lung cancer patients will suffer from brain metastases (BM) (1,2). MicroRNA (miRNA) is a small non-coding RNA with a length of 20–24 nucleotides, and plays an essential role in inhibiting messenger RNA (mRNA) translation and promoting mRNA degradation by base pairing to complementary sites of target mRNAs. Aberrant miRNA expression is closely associated with pathophysiological processes of various carcinomas and is a promising biomarker for the early diagnosis of lung cancer (3-5). Therefore, there is an urgent need to develop an accurate miRNA detection technique for the diagnosis of lung cancer with BM.
The unique features of miRNA, such as small size, low content in tissues and cells, and similar sequences of miRNA family members in particular, increase the difficulty in establishing miRNA detection methods. The most common miRNA detection methods, including polymerase chain reaction (PCR) (6-8), northern blotting (9,10), and microarray (11), have been widely applied in clinical sample analyses. Even though these methods have been the most common and gold standard methods, they are also criticized for their own shortcomings. For example, polyacrylamide gel electrophoresis (PAGE) separation which consists of transferring membrane and multiple washing procedures is essential, whereas complex in the northern blotting. Microarray assay has a high cost of production and detection, and the complicated probe design can lead to poor specificity and sensitivity in PCR-based miRNA detection. Over the past few years, a number of methods have been proposed due to their potential in the accurate detection of miRNA (12-17). For example, Zhao et al. proposed an intracellular miRNA detection method through integrating upconversion nanoparticles based on light trigger and catalytic hairpin assembly (CHA), and obtained an fM level sensitivity for miRNA detection (18). Wang et al. established accurate miRNA detection through integrating rolling circle amplification (RCA) for signal amplification and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nucleases 9 (Cas9) for signal generation (19,20). There is increased demand for efficient techniques and other alternative approaches with more accurate sensitivity, reasonable cost, and concise detection. Therefore, there is a need to develop a sensitive, accurate, and user-friendly method for more efficient miRNA detection. In the past few years, because of the requirements of amplification effectiveness and constant temperature operation, isothermal signal amplification techniques have been used for both in vitro and in situ miRNA detection and have been the focus of many researchers. Of these, the RCA technique has been demonstrated to be a suitable method to amplify miRNA in vitro and has also been applied to amplify RNA for in situ imaging or logic gates construction (2). According to RCA-based amplification, single-stranded nucleic acids (miRNA, short DNA) could be successfully transferred into long single-stranded DNA (ssDNA) products. However, RCA could only achieve limited amplification, and is therefore not the best choice for miRNA detection, because miRNA detection requires extremely high sensitivity.
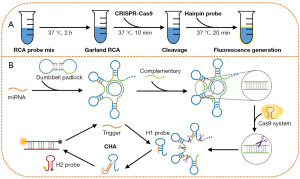
Therefore, in the present study, we developed the CRISPR/Cas9-assisted garland RCA approach for the precise detection of miRNA. In this method, target miRNA binds with 2 terminals of dumbbell padlock and cyclizes the padlock. With the assistance of T4 DNA ligase and phi29 DNA polymerase, an ssDNA product was obtained with repeated hairpin domains. After recognition with complementary sequences, a double-stranded DNA (dsDNA) section is formed that can be identified by the CRISPR/Cas9 system. After cleavage of the dsDNA section by the Cas9 enzyme, the RCA product is dissociated into hairpin probes to trigger the CHA-based signal amplification and generation. The proposed approach is promising for miRNA detection and lung cancer diagnosis. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1405/rc).
Methods
Materials
All the sequences used in this research were purchased from Sangon (Shanghai, China) (Table S1). Phi29 DNA polymerase, T4 DNA ligase, dNTPs, and bovine serum albumin (20 mg/mL) used for RCA were purchased from New England Biolabs (Ipswich, MA, USA). Diethyl pyrocarbonate water was purchased from Sigma Aldrich (St Louis, MO, USA). The Cas9 enzyme and its CRISPR RNA (crRNA) sequences were obtained from Inovogen (Inovogen, China). Fluorescence signal was detected using a Hitachi fluorescence spectrophotometer F-4700 (Hitachi, Japan). The Mini-PROTEAN vertical electrophoresis apparatus was purchased from Bio-Rad (Bio-Rad, Hercules, CA, USA).
RCA fluorescence analysis
The RCA process was performed according to the following steps: 2 µL of the padlock template, 2 µL miRNA-21 (100 pM each), 1 µL T4 DNA ligase (2,000 U/mL), 1 µL phi29 DNA polymerase (10 U/µL), and 8 µL dNTP mixture were first added in a reaction buffer containing 50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, and 2 µL Dithiothreitol (10 mM, pH 7.5) at 25 ℃. The mixture was then incubated at 37 ℃ for 1 h, slightly vortexed, and spun down. Afterwards, the mixture was heated to 90 ℃ for 5 min and gradually cooled to room temperature. In total, 2 µL Cas9 enzyme (1 U/L) and 2 µL synthesized RNA sequences were added in the mixture and incubated at 37 ℃ for 20 min; 2 µL H2 probe (1 nM) was then added to the mixture. The fluorescence signal of the whole mixture was detected when it was incubated with the H2 probe for 20 min.
Statistical analysis
Statistical significances of all collected data were calculated by GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA), and all data were expressed as mean ± standard deviations. Two-tailed Student’s t-test was used to compare differences between two groups, with P<0.01 indicating statistical significance.
Results
Working mechanism of the proposed method
Detection procedures of the proposed method are illustrated in Figure 1A. In the RCA process, target miRNA could hybridize with 2 terminals of the designed dumbbell padlock to form a cyclized padlock with the assistance of the T4 DNA ligase. With the assistance of phi29 DNA polymerase for chain extension, a garland RCA product was obtained with a repeated hairpin structure section (Figure 1B). A dsDNA section was formed that could be recognized by the CRISPR/Cas9 system with the addition of the complementary sequences. Based on the CRISPR/Cas9-assisted specific cleavage of the formed dsDNA section, the garland RCA product was broken into hairpin structure probes (H1 probe). The generated hairpin probes could be unfolded by targeted miRNA and formed the miRNA-H1 probe complex. Fluorescence-labeled hairpin probe-2 (H2 probe) was added to hydride with the other terminal of H1 probe and replace trigger, liberated to trigger and attend next recycle which composed the CHA amplification process.
Feasibility of the method
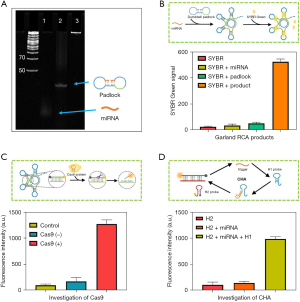
PAGE gel electrophoresis was used to determine the feasibility of RCA in this method. As shown in Figure 2A, miRNA (lane 1) and padlock (lane 2) moved faster than their hybrid products, while RCA products were blocked in the loading hole, indicating the success of RCA. The formation of the hairpin structure in the RCA product was analyzed by fluorescence experiment. In the assembly of the garland RCA product, the obtained RCA products were heated to 90 ℃ for 10 min and then slowly brought to room temperature. The formation of the hairpin structure could be seen through the formation of the stem section. SYBR Green (Solarbio, Beijing, China) was used to label the stem section. As shown in Figure 2B, the obtained signal was much higher when the garland RCA product existed compared with only SYBR Green, indicating the success of the garland RCA product. We then investigated whether CRISPR/Cas9 could specifically recognize and cut the formed dsDNA section in garland RCA products by adding complementary sequences, which were labeled with fluorescein groups and quenched groups at both ends. The results showed that the fluorescence signal in the system was significantly enhanced in the presence of Cas9, suggesting that the complementary sequence was cut apart, leading to the re-emergence of Cy3 fluorescence (Figure 2C). We then studied the CHA process by synthesized H1 probe and H2 probe. As seen in Figure 2D, the obtained fluorescence signal of the H2 probe showed time-dependent enhancement, suggesting the feasibility of the CHA process.
Optimization of experimental conditions
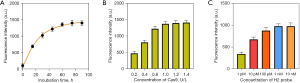
We then optimized the related experimental conditions to achieve superior detection performance. Considering the importance of RCA in the overall approach, we investigated RCA amplification time, T4, and phi29 enzyme concentration. As shown in Figure 3A, the obtained fluorescence intensity increased with the increase of incubation time and did not increase significantly after more than 60 min of incubation. The T4 and Phi29 enzymes were finally optimized to 1 and 1.5 U/L, respectively (Figure S1). We then verified the concentration of then Cas9 enzyme. As shown in Figure 3B, when the concentration of Cas9 increased, the detected signal increased correspondingly, but when the concentration exceeded 1 U/L, the fluorescence signal did not change significantly. The optimized H2 concentration for miRNA detection in the CHA process was tested. As shown in Figure 3C, the obtained florescence intensity of the system increased when the H2 probe concentration varied from 1 pM to 1 nM, and there were no furthers increases of florescence at concentrations >1 nM.
Detection performance of the method
Under optimal experimental conditions, the sensitivity of this method was determined by detecting miRNA synthesized under different concentration gradients (10 fM to 100 pM). As shown in Figure 4A, the fluorescence intensity at 570 nm gradually increased with the concentration in the range of 10 fM to 100 pM. In addition, the data were fitted by exponential curve to obtain the following fluorescence concentration curve: Y=312.3 × lgC + 2108 (R2=0.9786, where C is the concentration of target miRNA) (Figure 4B). the limit of detection of the established approach was determined 3.45 fM according to the general 3σ method. These findings indicate that RCA has high detection sensitivity for the amplification of miRNA signals. The specificity of the method in 1× phosphate buffered saline buffer and clinical serum samples was evaluated. As shows in Figure 4C, the fluorescence intensity obtained of the target miRNA group was significantly higher than average level that obtained by other miRNA groups. The results demonstrated high selectivity of the established approach in identifying target miRNA from homologous ones.

Clinical application of the method
To investigate the potential clinical application of the constructed CRISPR/Cas9-assisted RCA system for miRNA detection, we applied this protocol to detect miRNA in serum samples from lung cancer patients with and without BM. Studies have reported that miRNA-326 is an anti-oncogene that inhibits tumor metastasis, so it is considered a potential biomarker for lung cancer detection (21). As shown in Figure 5, the concentration of target miRNA obtained by lung cancer patients with BM was much lower than that obtained by lung cancer patients without BM, which was consistent with previous reports (22).

Discussion
We established an accurate and sensitive miRNA detection technique through the CRISPR/Cas9-assisted integration of garland RCA and CHA. In this method, target miRNA could first be amplified into garland RCA products, which contain a complementary sequence recognizing section. With the addition of a complementary sequence, dsDNA was formed that could be recognized and cut by the Cas9 enzyme, generating a large amount of H1 probes to induce CHA-based signal amplification. Eventually, the established approach possessed a wide detection range from 10 fM to 100 pM with the LOD as low as 3.45 fM, which is comparable to most of the former established miRNA detection approaches. In addition, the method also exhibited a high selectivity to target miRNA assured by the high specificity of CRISPR-Cas9 system. Several high highlights of the approach could be summarized as: (I) the padlock sequences were designed with dumbbell structure to reduce the possible mismatch with target miRNA, improving the detection specificity; (II) RCA and CHA process were integrated in the established approach by the CRISPR-Cas9 based chain specific cleavage, endowing the method a high sensitivity; (III) CRISPR-Cas9 system specifically identify and cut the complemented section, checking the RCA products and adding the specificity of the approach. Despite all the above advantages, the requirement of labeling the FAM moiety inevitably increased the background signals. in the future, we will focus on developing the novel biosensors that can detect miRNA in a label-free way. Our proposed method for miRNA detection could be used in the screening, diagnosis of lung cancer with BM. In the future, we will focus on improving the sensitivity of our approach for clinical use.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant No. 81672959).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1405/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1405/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1405/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lareau S, Slatore C, Smyth R. Lung Cancer. Am J Respir Crit Care Med 2021;204:21-2. [Crossref] [PubMed]
- Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol 2021;33:40-6. [Crossref] [PubMed]
- Bica-Pop C, Cojocneanu-Petric R, Magdo L, et al. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci 2018;75:3539-51. [Crossref] [PubMed]
- I H. Cho JY. Lung Cancer Biomarkers. Adv Clin Chem 2015;72:107-70. [Crossref] [PubMed]
- Iqbal MA, Arora S, Prakasam G, et al. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med 2019;70:3-20. [Crossref] [PubMed]
- Jet T, Gines G, Rondelez Y, et al. Advances in multiplexed techniques for the detection and quantification of microRNAs. Chem Soc Rev 2021;50:4141-61. [Crossref] [PubMed]
- Forero DA, González-Giraldo Y, Castro-Vega LJ, et al. qPCR-based methods for expression analysis of miRNAs. Biotechniques 2019;67:192-9. [Crossref] [PubMed]
- Takei F, Akiyama M, Murata A, et al. RT-Hpro-PCR: A MicroRNA Detection System Using a Primer with a DNA Tag. Chembiochem 2020;21:477-80. [Crossref] [PubMed]
- Usó M, Jantus-Lewintre E, Sirera R, et al. miRNA detection methods and clinical implications in lung cancer. Future Oncol 2014;10:2279-92. [Crossref] [PubMed]
- Ahmad W, Gull B, Baby J, et al. A Comprehensive Analysis of Northern versus Liquid Hybridization Assays for mRNAs, Small RNAs, and miRNAs Using a Non-Radiolabeled Approach. Curr Issues Mol Biol 2021;43:457-84. [Crossref] [PubMed]
- Jin F, Xu D. A fluorescent microarray platform based on catalytic hairpin assembly for MicroRNAs detection. Anal Chim Acta 2021;1173:338666. [Crossref] [PubMed]
- Voss G, Edsjö A, Bjartell A, et al. Quantification of microRNA editing using two-tailed RT-qPCR for improved biomarker discovery. RNA 2021;27:1412-24. [Crossref] [PubMed]
- Koo KM, Carrascosa LG, Shiddiky MJ, et al. Poly(A) Extensions of miRNAs for Amplification-Free Electrochemical Detection on Screen-Printed Gold Electrodes. Anal Chem 2016;88:2000-5. [Crossref] [PubMed]
- Li J, Liu J, Bi Y, et al. Ultrasensitive electrochemiluminescence biosensing platform for miRNA-21 and MUC1 detection based on dual catalytic hairpin assembly. Anal Chim Acta 2020;1105:87-94. [Crossref] [PubMed]
- Lu W, Wang Y, Song S, et al. A fishhook probe-based rolling circle amplification (FP-RCA) assay for efficient isolation and detection of microRNA without total RNA extraction. Analyst 2018;143:5046-53. [Crossref] [PubMed]
- Wang F, Wang H, Zhang P, et al. Ultrasensitive multiplexed detection of miRNA targets of interest based on encoding probe extension in improved cDNA library. Anal Chim Acta 2021;1152:338281. [Crossref] [PubMed]
- Xu S, Chang Y, Wu Z, et al. One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens Bioelectron 2020;149:111848. [Crossref] [PubMed]
- Zhao X, Zhang L, Gao W, et al. Spatiotemporally Controllable MicroRNA Imaging in Living Cells via a Near-Infrared Light-Activated Nanoprobe. ACS Appl Mater Interfaces 2020;12:35958-66. [Crossref] [PubMed]
- Wang R, Zhao X, Chen X, et al. Rolling Circular Amplification (RCA)-Assisted CRISPR/Cas9 Cleavage (RACE) for Highly Specific Detection of Multiple Extracellular Vesicle MicroRNAs. Anal Chem 2020;92:2176-85. [Crossref] [PubMed]
- Qiu XY, Zhu LY, Zhu CS, et al. Highly Effective and Low-Cost MicroRNA Detection with CRISPR-Cas9. ACS Synth Biol 2018;7:807-13. [Crossref] [PubMed]
- Pan YJ, Wan J, Wang CB. MiR-326: Promising Biomarker for Cancer. Cancer Manag Res 2019;11:10411-8. [Crossref] [PubMed]
- Shao L, He Q, Wang J, et al. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discov 2021;7:145. [Crossref] [PubMed]
(English Language Editor: R. Scott)


