
Laboratory blood test profiling reveals distinct biochemical and hemocyte features of KRAS mutated non-small cell lung cancer
Highlight box
Key findings
• This study analyzed the laboratory blood indexes of non-small cell lung cancer (NSCLC) patients with different genotypes and found that the biochemical and hemocyte features of KRAS mutated patients were significant differences.
What is known and what is new?
• NSCLC of different driver gene subgroups have substantial molecular and clinical heterogeneities;
• The study revealed that different gene types can lead to differences in metabolic levels, which are expressed in the blood test indicators of patients. In response to the findings, a non-invasive and more cost-effective prognostic model was developed using the laboratory parameters for predicting KRAS mutations.
What is the implication, and what should change now?
• The model can be used as a practical screening tool to guide the use of more specialized and expensive molecular assays for KRAS mutation in NSCLC.
Introduction
Lung cancer is the most commonly diagnosed cancer worldwide and the leading cause of cancer death. Its mortality rate is higher than that of colorectal cancer, stomach cancer, and liver cancer (1). It is a heterogeneous disease in which disease progression is affected by genetic factors and environmental exposure. The treatment of lung cancer depends on factors such as histological type, tumor stage, genotype, lung function, and patient status. Non-small cell lung cancer (NSCLC) shows distinct genetic drivers and divergent prognostic profiles. Therapeutic clinical trials in NSCLCs indicate differential different subtypes response to treatments. Hence, NSCLC appear to be vastly distinct diseases at the molecular, pathological, and clinical level (2,3). About half of patients with NSCLC have the genetic mutation and are eligible for tyrosine kinase inhibitor (TKI) targeted therapy (4). Improved understanding of the biology and molecular subtypes of NSCLC have led to more biomarker-directed therapies for patients. These biomarker-directed therapies and newer empirical treatment regimens have improved overall survival (5,6) and higher health utility value for patients with NSCLC (7,8).
Similarly, tumor heterogeneity, biological behavior, and patient physical condition are associated with histological subtypes and types of genetic mutations. Previous studies have suggested that there are substantial molecular and clinical heterogeneities in different driver gene subgroups (9,10). Metabolic dysregulation is an important mechanism for tumorigenesis and development, with mutations in some oncogenes and/or tumor suppressor genes that can mediate the metabolic reconnection of cancer cells to support these cells’ high requirements for building blocks and energy production. Since cancer cells are prone to multiple oncogenic mutations, such as RAS, EGFR, MYC, and BRAF mutations, these genes may also affect the metabolic changes of cancer (11-13), including glucose, lipid, amino acid, and nucleic acid metabolism (14). For example, different MYC-driven small-cell lung cancer subtypes have been reported to have distinct metabolic profiles, and they are preferentially dependent on arginine-regulated pathways (15). KRAS mutations are associated with glycolysis, enhanced glutathione-mediated detoxification (16), and enhanced oxidative phosphorylation in tumors (17).
Based on the above studies, it is reasonable to believe that different gene types can lead to differences in metabolic levels, which are expressed in the blood test indicators of patients. To validate this hypothesis, we analyzed 46 laboratory variables for lung cancer patients with five gene types and observed and analyzed the differences between them. On this basis, we constructed a simple predictive model using important variables found in the least absolute shrinkage and selection operator (LASSO) regression to predict the KRAS mutation of patients, which can help doctors assess the patient’s genetic mutation status as early as possible before surgery. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-829/rc).
Methods
Study population
The hematological parameters of 1,688 patients diagnosed with lung cancer and underwent surgical resection from the First Affiliated Hospital of Guangzhou Medical University from February 2010 to July 2018 were retrospectively evaluated. All samples of primary surgery patients who had not undergone anti-tumor therapy were obtained from the last preoperative blood test. Ninety-three patients with abnormally high or low results were excluded. The following results were included for analysis: routine blood tests, biochemical tests, coagulation function tests, arterial blood gas analysis, lipid profile, renal function, electrolyte levels, serum tumor markers, and blood pressure. In addition, the last blood index results of 197 consecutive patients with NSCLC in the Cancer Center of Sun Yat-sen University from December 2015 to May 2016 and 147 patients diagnosed with colorectal cancer in our center at 2015 to 2016 were selected as verification cohorts. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was confirmed and approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. 2015-25). Since the study was a retrospective analysis of the patient database, the requirement for informed consent of each patient was waived.
Data collection
Demographic variables collected for the study included age, sex, smoking status, and medical history. Basic clinical variables included tumor stage, site, size, pathological type, and genotype (Table 1). A total of 46 laboratory test index variables were included in the study, including: white blood cell count (WBC), red blood cell count (RBC), platelet (PLT), hemoglobin (Hb), neutrophil count (NEUT), lymphocyte count (LYM), monocyte count (MONO), eosinophil count (EOS), basophil count (BASO), prothrombin time (PT), activate part plasma prothrombin time (APTT), fibrinogen (FIB), D-dimer, international normalized ratio (INR), arterial oxygen partial pressure (PaO), arterial blood carbon dioxide partial pressure (PaCO2), arterial oxygen saturation (SaO), pH, base excess (BE), standard bicarbonate (SB), uric acid (UA), α-hydroxybutyrate dehydrogenase (α-HBDH), total cholesterol (TC), triglycerides (TG), low-density lipoprotein cholesterol (LDL), high-density lipoprotein cholesterol (HDL), creatinine (Cr), blood urea nitrogen (BUN), blood sugar (GLU), serum potassium ion (K), sodium ion (Na), serum chlorine (CL), serum calcium (Ca), carbon dioxide combining power (CO2CP), aspartate aminotransferase (AST), serum creatine kinase (CK), serum creatine kinase isoenzyme (CK-MB), lactate dehydrogenase (LDH), alpha-L-fucosidase (AFU), carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CYFRA21-1), neuron specific enolase (NSE), carbohydrate antigen 125 (CA125), carbohydrate antigen 153 (CA153), and systolic and diastolic blood pressure.
Table 1
| Patient characteristic | Training set (n=1,595), n [%] | Testing set (n=197), n [%] | P value |
|---|---|---|---|
| Age, mean [range] (years) | 60 [18–88] | 56 [27–85] | <0.01 |
| Gender | 0.05 | ||
| Male | 849 [53] | 119 [60] | |
| Female | 746 [47] | 78 [40] | |
| Smoking | <0.01 | ||
| Never | 1,043 [65] | 105 [53] | |
| Current | 284 [18] | 54 [28] | |
| Former | 122 [8] | 28 [14] | |
| Unknown | 146 [9] | 10 [5] | |
| Tumor stage | <0.01 | ||
| IA | 803 [50] | 57 [29] | |
| IB | 258 [16] | 28 [14] | |
| IIA | 99 [6] | 13 [7] | |
| IIB | 109 [7] | 9 [5] | |
| IIIA | 210 [13] | 31 [16] | |
| IIIB | 42 [3] | 14 [7] | |
| IV | 74 [5] | 45 [22] | |
| Primary tumor site | <0.01 | ||
| Upper lobe | 910 [57] | 93 [47] | |
| Middle lobe | 107 [7] | 18 [9] | |
| Lower lobe | 494 [31] | 49 [25] | |
| Overlapping | 84 [5] | 37 [19] | |
| Tumor histology | 0.18 | ||
| Adenocarcinoma | 1,356 [85] | 158 [80] | |
| Squamous cell carcinoma | 142 [9] | 24 [12] | |
| Lymphoid epithelioid carcinoma | 30 [2] | 7 [4] | |
| Others | 67 [4] | 8 [4] |
Statistical analysis
Univariate analysis of candidate variables for multivariate LASSO regression screening
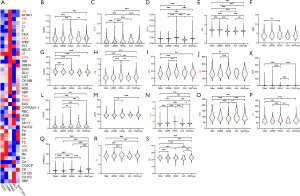
In this study, GraphPad Prism 8 and R software package were used for data analysis. In the training set, the median method filled in missing values because most continuous variables were not normally distributed. Heat maps were established using the median to demonstrate the relationship between blood test parameters in different groups. Univariate analysis was performed using the Kruskal-Wallis method to assess the associations between each index in different gene mutations. The results were described as violin maps in the quartile range (meaningful indicators are shown in Figure 1), observing the distribution and removing outliers. P value of less than 0.05 was considered statistically significant. Univariate analysis of 5 gene mutations was performed, and KRAS was observed as having the most significant difference. Therefore, KRAS was used as the outcome indicator for the next model.
LASSO regression and model construction
A strong multicollinearity relationship was observed since the 46 laboratory variables of a sample were derived from the same patient. All indicators for univariate analysis were introduced into the penalized logistic model to select variables for constructing a prediction model. LASSO was used for regression analysis of high-dimensional predictors. The approach has been extended and broadly applied to filter the variables to minimize the potential collinearity and confusion caused by overfitting. The R package “glmnet” was used to perform the LASSO regression. Meaningful variables screened by LASSO regression analysis were put into a logistic regression, and the prediction model was constructed with the coefficients weighted by the logistic model in the training cohort. A nomogram was developed according to the logistic model, which was internally validated using the bootstrap resampling method. The odds ratio (OR) and 95% confidence interval (CI) of the predicted variables were calculated by R package “glmnet”.
Model validation
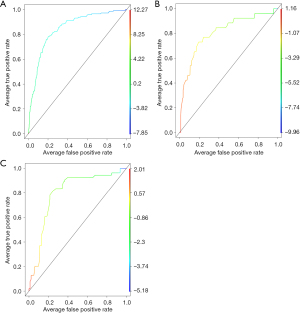
The calibration of the nomogram was carried out by internal validation using the bootstrap resampling approach, and the resulting nomogram was displayed using a calibration curve. The area under the receiver operating characteristic (ROC) curve (AUC) was calculated to measure the accuracy of the model. The clinical benefit of this nomogram model was further verified using the decision curve analysis. ROC curve was analyzed with R software and the “ROCR” package. Furthermore, 197 patients with pathologically diagnosed lung cancer from 2015 to May 2016 in the Sun Yat-sen University Cancer Center were randomly included as an independent verification cohort, whose gene types included EGFR19del, EGFR L858R, KRAS, ALK fusion, and Wildtype. The ROC curves of the training and verification groups were plotted in Figure 2 to show the performance of the model visually.
Results
Patient characteristics
Our analysis included a total of 1,792 NSCLC patients, who were unevenly divided into training groups (n=1,595) and verification groups (n=197) as described in the “ Methods” section. The numbers of KRAS mutation patients were 147 (9%) and 25 (13%) in the training set and validation set respectively. Table 1 summarizes the demographic and basic clinical variables in the training set and verification set. There was no significant difference in the distribution of these variables between them (P>0.05). Missing values in the original dataset were done by the median fill method. Eighteen of the 46 variables (MONO, EOS, BASO, PaO, TG, CO2CP, AFU, Cr, K, BUN, UA, HDL, APTT, FIB, CA125, CEA, CYFRA21-1, SB) showed a critical difference in the genotyping of each set of training sets.
Association between blood test results and driver mutation types
A heatmap was established using the median to preliminarily describe the relationship between blood indicators in different groups. KRAS mutation was observed to be a special type, and some metabolization-related indicators (such as TG, UA, etc.) showed significant differences (Figure 1A). Normalization analysis was performed on laboratory variables of each group of gene types, and Hb conforming to the normal distribution was analyzed using one-way analysis of variance (ANOVA). The remaining parameters did not conform to the normal distribution, and the Kruskal-Wallis test was used for the different analyses. Of the 46 laboratory variables, 18 showed significant inter-group differences in variance analysis (P<0.05) (Figure 1B-1S), including MONO, EOS, and BASO in blood routines, PaO, SB, and CO2CP in blood gas analysis, AFU, K, Cr, BUN, UA, TG and HDL in the biochemical examination, APTT and FIB in coagulation function, CA125, CEA and CYFRA21-1 in tumor markers. Among the 18 variables, the laboratory indicators of KRAS mutations were significantly different from the other four groups of genes, while the difference was not significant among the other four groups (Figure 1). Therefore, KRAS mutation was used as the outcome variable of the prediction model.
Variable screening
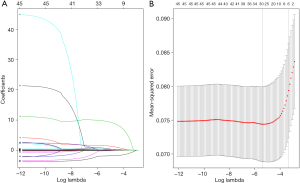
To adjust for potential confounding factors, we used the LASSO algorithm to select the most relevant features from the laboratory variables, which were then added to the Logistic model. The predictive correlation factor was selected by the minimum value of lambda (λ=0.0056) in LASSO (Figure 3), and included the following 28 variables: RBC, Hb, MONO, EOS, BASO, APTT, FIB, INR, PaO, SaO, BE, SB, UA, TG, LDL, HDL, Cr, GLU, K, Na, CO2CP, AST, CK, CK-MB, NSE, AFU, systolic and diastolic blood pressure.
Model construction and external verification
We further screened the significant variables in the LASSO-logistic regression model. Nine potential predictors were indicated in the training cohort and are presented in a nomogram plot (R2=0.257, C-index =0.85, Table 2), with non-zero coefficients as the features in the model (Table 2). We calculated the KRAS mutation as a diagnostic feature as follows: probability of KRAS mutation = (TG × 0.491) + (PaO × −0.034) + (UA × 0.007) + (BASO × 13.8) + (FIB × 0.352) + (SB × −0.159) + (HDL × −1.723) + (AFU × 0.070) + (EOS × 1.816). To confirm that the formula has a similar predictive value in different populations, the same formula was used in both internal verification and external verification cohorts. Consistent with the results of the training cohort, the ROC curve confirmed that the formula has good sensitivity and specificity in predicting KRAS mutations (the AUC of the training cohort and the validation cohort were 0.85 (95% CI: 0.81–0.88) and 0.81 (95% CI: 0.71–0.91) (Figure 2A,2B), respectively.
Table 2
| Variables | Unit | Coef | SE | Wald | Odds ratio | 95% CI | P value | |
|---|---|---|---|---|---|---|---|---|
| Lower limit | Upper limit | |||||||
| TG | mmol/L | 0.491 | 0.144 | 11.66 | 1.63 | 1.23 | 2.17 | 0.0006 |
| PaO | mmHg | −0.034 | 0.006 | 28.32 | 0.97 | 0.96 | 0.98 | <0.0001 |
| UA | μmol/L | 0.007 | 0.001 | 25.7 | 1.01 | 1.00 | 1.01 | <0.0001 |
| BASO | 109/L | 13.828 | 3.496 | 15.65 | 1.41 | 1.07 | 1.12 | <0.0001 |
| FIB | g/L | 0.352 | 0.089 | 15..62 | 1.42 | 1.19 | 1.70 | <0.0001 |
| SB | mmol/L | −0.159 | 0.064 | 6.14 | 0.85 | 0.75 | 0.97 | 0.0132 |
| HDL | mmol/L | −1.723 | 0.616 | 7.84 | 0.18 | 0.05 | 0.59 | 0.0051 |
| AFU | U/g | 0.07 | 0.029 | 6.02 | 1.07 | 1.01 | 1.14 | 0.0142 |
| EOS | 109/L | 1.816 | 0.799 | 5.16 | 1.146 | 1.28 | 2.45 | 0.0231 |
Coef, coefficient; SE, standard error; CI, confidence interval; TG, triglycerides; PaO, arterial oxygen partial pressure; UA, uric acid; BASO, basophil count; FIB, fibrinogen; SB, standard bicarbonate; HDL, high-density lipoprotein cholesterol; AFU, alpha-L-fucosidase; EOS, eosinophil count.
Exploration of model predictive value in colorectal cancer
In the past decade, little attention has been paid to the relationship between KRAS mutant colorectal cancer and metabolic factors; only one study reported a specific association between KRAS mutant colorectal cancer and low plasma adiponectin levels (18). Therefore, we tried to apply this model to patients diagnosed with colorectal cancer in our center (2015–2016, n=147). Surprisingly, this diagnostic model also showed similar efficiency for those with colorectal cancer (R2=0.274, C-index =0.80, 95% CI: 0.72–0.88, Figure 2C). Upon further evaluation of blood test parameters, there was no significant difference between the different subtypes of KRAS mutation in the lung or colorectal cancer. A similar study by Cao et al. in 2020 showed that hematological parameters [WBC, MONO, monocyte/lymphocyte ratio (MLR), hematocrit (HCT), hemoglobin (HGB), mean platelet volume (MPV), mean corpuscular hemoglobin (MCH), mean corpusular hemoglobin concerntration (MCHC)] were significantly associated with KRAS gene mutations in colorectal cancer (19). Similar to our conclusions, the values of hematological parameters in patients with KRAS mutation were lower than in wild-type patients. The widespread distribution of the KRAS gene might contribute to worse physical conditions in patients with KRAS mutations than in wild-type patients through multiple pathways. However, metabolism-related parameters were not included in the study. Therefore, our study provided new evidence that KRAS gene mutations might specifically affect human metabolic processes.
Discussion
Over the past decade, great progress has been made in identifying gene mutations associated with various histological subtypes of lung cancer. The classification of lung cancer has evolved from a single disease to types of diseases with multiple histological subtypes and different molecular mutations. The National Comprehensive Cancer Network (NCCN) guidelines recommend that common mutation targets should be listed as routine detection items to guide effective targeted therapies (20). Genomics-based precision medicine has greatly improved patient survival, and determining genotyping plays a very important role in the treatment of lung cancer (21). However, some patients cannot obtain the lesion through biopsy or surgery. Next-generation sequencing (NGS) or multiplex PCR for multiple gene testing through blood tests is costly or even less effective. Furthermore, it is a huge burden for some non-wealthy patients to undergo multi-gene testing (22). Single-point testing can also be used, but other than the common mutations with treatment available, it is necessary to detect other genes in specific populations, such as KRAS (23,24).
EGFR 19del and 21 exon L858R mutations were previously considered to be a class of disease, but it had been confirmed that there were large differences between them with more and more recent data. Firstly, they had distinct molecular structures. Their kinase domain spatial configuration was located in different regions, which might affect the phosphorylation, leading to a discrepancy in subsequent functions. Secondly, their clinical characteristics were different. Patients with 19del were more dependent on the EGFR pathway and were significantly more sensitive to the drug than those with the 21 exon L858R mutation. In addition, the L858R mutation was associated with other mutations genes that might lead to bypass activation. Therefore, EGFR 19del and EGFR L858R mutations might have diverse biological characteristics. In this study, EGFR 19del and EGFR L858R were considered independent factors of genotype. We compared the differences between 46 laboratory variables that must be routinely tested before surgery for routine clinical monitoring in five gene types and found significant differences for 18 variables in NSCLC, especially in patients with KRAS mutations. The values of the metabolic-related parameters (such as TG, HDL, UA, etc.) in patients with KRAS mutation showed more deviation from the normal values than the remaining four mutant lung cancer patients. Some parameters, such as BASO and EOS, may not be related to cell metabolism. We considered that the abnormality of these parameters was associated with immuno-inflammatory phenotypes and immunogenic enhancements in KRAS mutant lung cancer.
KRAS is one of the first oncogenes found to be mutated in human cancers, including lung, colorectal and pancreatic cancers. Tumors driven by mutant KRAS are the most invasive and refractory. Most KRAS mutations occur at codons 12 and 13 (25). All three common G12 mutations (G12C, G12V, and G12R) were associated with poor prognosis (26). All KRAS mutations are believed to lead to tumor development and growth by activating a series of complex downstream signaling pathways, including mitogen-activated protein kinases, but no effective drug had been developed to suppress this mechanism (25). Recently, however, some studies have discovered the role of this carcinogenic mutation in reconnecting the cancer cell metabolism, and new therapeutic opportunities have emerged. Cancer cells that rely on KRAS-driven metabolic adaptation are very sensitive to the inhibition of these metabolic pathways, thus revealing new interventional therapeutic windows. Generally, the mutant KRAS promotes tumor growth by transferring the metabolism of cancer cells toward the anabolic pathway. In tumor, KRAS-driven metabolic rearrangement is performed by up-regulating rate-limiting enzymes involved in amino acid, fatty acid, or nucleotide biosynthesis and stimulating clearance pathways (such as macrophage proliferation and autophagy), which in turn provides the basic anabolic pathway (27). However, these studies were conducted only at the cellular or animal level. Our study further validates the effect of KRAS mutations on human metabolic pathways from a clinical perspective.
In response to this important finding, we developed a simple and more cost-effective prognostic model using the aforementioned laboratory parameters to help physicians assess the KRAS mutation status as early as possible before surgery, providing a basis for precise medication and early treatment. Our model uses laboratory indicators (including blood routine, biochemical, coagulation, and blood pressure levels) that should be monitored in each patient before surgery, which are readily available and relatively inexpensive. With an AUC of 0.85 (95% CI: 0.81–0.88) via internal validation and the use of the bootstrap resampling method, our model exhibited a sufficient ability to predict the KRAS mutation status. We also used a cohort from Sun Yat-sen University Cancer Center to conduct external model validation with an AUC of 0.81 (95% CI: 0.71–0.91). The calibration curve further demonstrates the effectiveness of the prediction performance. The decision curve analyses suggest that the nomogram model could obtain a favorable net benefit when the risk threshold was less than 0.6, which may be related to the low mutation rate of KRAS in Asian people. If the prediction probability of the model is high, some false positives might occur. The results show that the model has high prediction accuracy and reliability, and even the possibility for application in multiple cancer types. In addition, blood tests routinely used in clinical medicine are more reliable than most tests performed in biological laboratories and do not require specialized equipment or expertise.
Our study has several limitations. Firstly, the dates of patients’ diagnoses are between 2010 and 2018. This is a relatively long period and may increase the heterogeneity of our population, especially in terms of treatment and testing. We included a sample size large enough to counteract the bias caused by individual differences as much as possible. Nevertheless, this heterogeneity may increase the versatility of the model. Secondly, due to the inconsistency of preoperative monitoring indicators, some indicators have more missing values than others. Although the actual observations are estimated by the median fill method, it is still unclear whether the performance of the model can be further improved by including more clinical and laboratory variables and a more complete data set. Thirdly, the study used a retrospective icon review rather than a prospective clinical trial. These facts inevitably limit the clinical utility of current research.
In summary, we developed a non-invasive and less costly screening model using a hospital-based multicenter research cohort based on objective data that is readily available, which demonstrates a high predictive performance of KRAS mutation status in lung cancer patients.
Acknowledgments
We would like to thank Lindsey Hamblin for her assistance in language editing our paper. We appreciate the coordination of all the patients who contributed to this study. The abstract of this study was displayed as a poster at the European Lung Cancer Conference (ELCC 2020), which was entitled Predicting KRAS mutation of NSCLC based on laboratory blood test profiling. Thanks for the exhibition opportunity given by the conference officials.
Funding: This study was supported by the National Natural Science Foundation of China (grant No. 81772486); the Natural Science Foundation for Distinguished Young Scholars of Guangdong Province (grant No. 2014A030306013).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-829/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-829/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-829/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-829/coif). WL serves as an unpaid editorial board member of Journal of Thoracic Disease. WL reports that this study was supported by the National Natural Science Foundation of China (grant No. 81772486); the Natural Science Foundation for Distinguished Young Scholars of Guangdong Province (grant No. 2014A030306013). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was confirmed and approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. 2015-25). Since the study was a retrospective analysis of the patient database, the requirement for informed consent of each patient was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Relli V, Trerotola M, Guerra E, et al. Abandoning the Notion of Non-Small Cell Lung Cancer. Trends Mol Med 2019;25:585-94. [Crossref] [PubMed]
- Sacher AG, Dahlberg SE, Heng J, et al. Association Between Younger Age and Targetable Genomic Alterations and Prognosis in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:313-20. [Crossref] [PubMed]
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637-58. [Crossref] [PubMed]
- Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019;322:764-74. [Crossref] [PubMed]
- Schulze AB, Evers G, Kerkhoff A, et al. Future Options of Molecular-Targeted Therapy in Small Cell Lung Cancer. Cancers (Basel) 2019;11:690. [Crossref] [PubMed]
- Yang SC, Kuo CW, Lai WW, et al. Dynamic Changes of Health Utility in Lung Cancer Patients Receiving Different Treatments: A 7-Year Follow-up. J Thorac Oncol 2019;14:1892-900. [Crossref] [PubMed]
- Singal G, Miller PG, Agarwala V, et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA 2019;321:1391-9. [Crossref] [PubMed]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Chen PH, Cai L, Huffman K, et al. Metabolic Diversity in Human Non-Small Cell Lung Cancer Cells. Mol Cell 2019;76:838-851.e5. [Crossref] [PubMed]
- Sellers K, Allen TD, Bousamra M 2nd, et al. Metabolic reprogramming and Notch activity distinguish between non-small cell lung cancer subtypes. Br J Cancer 2019;121:51-64. [Crossref] [PubMed]
- Visweswaran M, Arfuso F, Warrier S, et al. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells 2020;38:6-14. [Crossref] [PubMed]
- Hensley CT, Faubert B, Yuan Q, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016;164:681-94. [Crossref] [PubMed]
- Chalishazar MD, Wait SJ, Huang F, et al. MYC-Driven Small-Cell Lung Cancer is Metabolically Distinct and Vulnerable to Arginine Depletion. Clin Cancer Res 2019;25:5107-21. [Crossref] [PubMed]
- Kerr EM, Gaude E, Turrell FK, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016;531:110-3. [Crossref] [PubMed]
- Caiola E, Falcetta F, Giordano S, et al. Co-occurring KRAS mutation/LKB1 loss in non-small cell lung cancer cells results in enhanced metabolic activity susceptible to caloric restriction: an in vitro integrated multilevel approach. J Exp Clin Cancer Res 2018;37:302. [Crossref] [PubMed]
- Inamura K, Song M, Jung S, et al. Prediagnosis Plasma Adiponectin in Relation to Colorectal Cancer Risk According to KRAS Mutation Status. J Natl Cancer Inst 2016;108:djv363. [Crossref] [PubMed]
- Cao Y, Gu J, Yan L, et al. The value of haematological parameters and serum tumour markers for predicting KRAS mutations in 784 Chinese colorectal cancer patients: a retrospective analysis. BMC Cancer 2020;20:1099. [Crossref] [PubMed]
- Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. [Crossref] [PubMed]
- Sousa AC, Silveira C, Janeiro A, et al. Detection of rare and novel EGFR mutations in NSCLC patients: Implications for treatment-decision. Lung Cancer 2020;139:35-40. [Crossref] [PubMed]
- Presley CJ, Tang D, Soulos PR, et al. Association of Broad-Based Genomic Sequencing With Survival Among Patients With Advanced Non-Small Cell Lung Cancer in the Community Oncology Setting. JAMA 2018;320:469-77. [Crossref] [PubMed]
- Guibert N, Barlesi F, Descourt R, et al. Characteristics and Outcomes of Patients with Lung Cancer Harboring Multiple Molecular Alterations: Results from the IFCT Study Biomarkers France. J Thorac Oncol 2017;12:963-73. [Crossref] [PubMed]
- Papke B, Der CJ. Drugging RAS: Know the enemy. Science 2017;355:1158-63. [Crossref] [PubMed]
- Murugan AK, Grieco M, Tsuchida N. RAS mutations in human cancers: Roles in precision medicine. Semin Cancer Biol 2019;59:23-35. [Crossref] [PubMed]
- Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol 2015;10:431-7. [Crossref] [PubMed]
- Kimmelman AC. Metabolic Dependencies in RAS-Driven Cancers. Clin Cancer Res 2015;21:1828-34. [Crossref] [PubMed]


