
YTHDC2-mediated m6A mRNA modification of Id3 suppresses cisplatin resistance in non-small cell lung cancer
Highlight box
Key findings
• RNA methyltransferase YTHDC2 might be a new target for cisplatin-resistant lung cancer treatment.
What is known and what is new?
• Id3 has the potential to reverse cisplatin resistance in lung adenocarcinomas.
• YTHDC2 inhibited the m6A level of Id3, which eventually inhibited NSCLC resistance to cisplatin.
What is the implication, and what should change now?
• We have identified a new tumor suppressor in NSCLC resistant to cisplatin called YTHDC2. The information will be useful for developing next-generation therapeutics.
Introduction
In China and worldwide, lung cancer is the most common cancer, and it accounts for the highest death rate among cancers (1). A major clinical problem is resistance to cisplatin therapy, which contributes to poor patient outcomes. Usually, changing oncogenes and tumor suppressor genes genetically and epigenetically cause lung cancer to develop in a multistage process (2). Epigenetic mechanisms for cisplatin resistance in lung cancer remain largely unexplored.
In lung adenocarcinomas, Id3 modulates PI3K/Akt to reverse cisplatin resistance (3). There is general agreement that Id3 plays a role in lung cancer proliferation and in resistance to cisplatin, but its exact mechanism remains unclear. Epigenetic regulation at a new level, the post-transcriptional level, is emerging through RNA modifications, in contrast to DNA methylation and histone modification (4). N6-methyladenosine (m6A) is the most common and abundant messenger RNA (mRNA) modification, modulated by ‘writers’, ‘erasers’, and ‘readers’ of this mark (5). The complex of writers, which includes METTL14, METTL3, and WTAP, promotes m6A modification in RNA. Ye et al. (6) found decreased IncRNA DBH-AS1 expression in pancreatic cancer was related to the METTL3-dependent m6A methylation of the lncRNA, which downregulation was negatively correlated with the malignancy of pancreatic cancer and with patient survival outcomes. Chen et al. (7) discovered the mechanism through which m6A methylation modulates autophagy and chemosensitivity in the TCam-2 cell line, and METTL3 was identified as a possible target to overcome seminoma cisplatin resistance. The mechanism of METTL3-mediated autophagy in reversing gefitinib resistance of non-small cell lung cancer (NSCLC) cells by β-elemene was revealed by Liu et al. (8). In contrast, erasers in m6A-modified mRNA counteract the effects of writers. The first identified demethylase was FTO. FTO might play an important role in promoting NSCLC by lowering m6A levels and activating KRAS signaling, as described by Shi et al. (9), who for the first time identified the genetic changes in m6A regulatory factors in NSCLC. The readers convert instructions of m6A modification into functional signals, including YTH family proteins. In the study by Tsuchiya et al. (10), as RNA-binding proteins(readers) that recognize m6A, high expressions of both YTHDF1 and YTHDF2 are connected to a favorable prognostic outcome of NSCLC patients, a larger quantity of tumor-infiltrating lymphocytes (TILs), and downregulation of programmed cell death 1 (PD-1).
Since the m6A modification machinery plays a crucial role in normal bioprocesses, it is not surprising that evidence is emerging that dysregulation of m6A modification and the associated proteins also contributes to cancer initiation, progression, and response to drugs (11). In recent studies, YTHDC2 was identified as an essential member of the m6A reading proteins involved in the development and progression of a variety of tumors. He et al. reported that as YTHDC2 activates the IGF1R/ATK/S6 signaling axis, it may serve as a potential therapeutic target in radiosensitization of neural progenitor cells (12). It was found that alterations in YTHDC2 were associated with better prognosis in head and neck squamous cell carcinoma (HNSCC), which may serve as a novel prognostic biomarker (13). However, the mechanism of m6A modification in the regulation of cisplatin resistance in lung cancer is unclear.
In this study, the downregulation of YTHDC2 expression was associated with a poor prognosis of lung cancer in cisplatin-resistant cells, which were expressed at a lower level than in sensitive cells. In addition, we demonstrated that YTHDC2, as a specific reader of m6A, inhibited the m6A level of Id3, which inhibited the proliferation and migration of A549/DDP cells and ultimately inhibited NSCLC resistance to cisplatin. As a result of our findings, we propose that the RNA methyltransferase YTHDC2 may serve as a new target for lung cancer cisplatin-resistant therapy. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-187/rc).
Methods
Clinical samples
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was reviewed and approved by the Ethics Committee of Jinling Hospital (No. 2020DZGZRZX-096). All participants signed an informed consent form. During June 2020 and September 2021, a total of 9 cases of NSCLC tissues were collected from patients in Jinling Hospital. Among them, 5 cases were in cisplatin-sensitive tissues and 4 cases were in cisplatin-resistant tissues. The following were the inclusion requirements: NSCLC was confirmed by a histopathological examination, and first-line chemotherapy included at least 3 cycles of a cisplatin-based combination therapy. A total of 9 patients enrolled in the study received cisplatin chemotherapy and were evaluated for sensitivity or resistance to cisplatin by computed tomography (CT) before and after cisplatin treatment. In addition, 3 cases of paracancerous tissues were also collected. All tissues were placed in liquid nitrogen for storage immediately after collection.
Cell lines, cell culture, vector construction, and transfection
In A549 (human species, KG007, KeyGen Biotech, Nanjing, China) cells, 10% fetal bovine serum (FBS) was added to Roswell Park Memorial Institute (RPMI)-1640 media, whereas in A549/DDP (human species, KG354, KeyGen Biotech) cells, 1,000 ng/mL DDP and 10% FBS were added to the RPMI-1640 media. After amplifying YTHDC2’s full-length coding sequence by polymerase chain reaction (PCR), and cloning it into pCDH vector, the recombinant plasmid was named pCDH-CMV-hYTHDC2(NM_022828.5)-C-3Flag-EF1A-GFP-T2A-Puro. Lipo8000™ transfection reagent was purchased from Beyotime Biotechnology (Shanghai, China). Transfection was carried out when approximately 80% confluency was reached in 96- or 6-well plates. For each transfection of 6-well plate, the following was added to each well: 2.5 ng plasmid DNA, 4 µL Lipo8000™ transfection reagent, and in 125 µL Opti-MEM (Gibco, Grand Island, NY, USA). After 24–48 h, cells were successfully transfected for subsequent analyses.
RNA isolation and quantitative real-time PCR (qPCR)
The RNA was isolated using TRIzol (Takara, Shiga, Japan), and its concentration was determined using a NanoDrop 2000. Using a reverse transcription kit, complementary DNA (cDNA) was created (RR036A; Takara). Using a TB Green Premix Extra kit, the real-time PCR (RT-PCR) reaction was performed (RR420A; Takara). Our qPCR reactions were conducted using Thermo Fisher Scientific’s Step One Plus Real-Time PCR System (Thermo Fisher, Waltham, MA, USA). We followed the manufacturer’s instructions for all subsequent steps. With glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the housekeeping gene, relative gene expression was calculated using the 2−ΔΔCt method. The primers used were as follows: GAPDH: forward 5'-CGCTCTCTGCTCCTCCTGTTC-3', revervse 5'-ATCCGTTGACTCCGACCTTCAC-3'; Id3 forward 5'-GGACGACATGAACCACTGCTACTC-3', reverse 5'-GCTGTAGGATTTCCACCTGGCTAAG-3'; YTHDC2 forward: 5'-CAAAACATGCTGTTAGGAGCCT-3', reverse 5'-CCACTTGTCTTGCTCATTTCCC-3'.
Western blot (WB) assay
The cells were treated with YTHDC2 or NC vector or YTHDC2 + m6A methylation inhibitor 3-deazaadenosine, after that, the cells were collected for WB. PBS was used to wash the cells twice. Proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer. A bicinchoninic acid (BCA) assay was performed on the supernatants after centrifugation to determine the concentration of protein. The protein concentration was determined using the standard curve. For electrophoresis, samples were prepared by adding 5× loading buffer and boiling for 5 min. Each protein was given a special polyacrylamide gel electrophoresis (PAGE) glue based on its molecular weight, and the samples were electrophoretically separated after adding the glue. Polyvinylidene fluoride (PVDF) membranes were applied to the electrophoresed products. After blocking the membrane for an hour at room temperature with 5% non-fat milk, the membrane was incubated with a primary antibody (Ab) then secondary Ab. Using electrochemiluminescence (ECL), we measured the intensity of protein expression. Antibodies used in this study were Id3 (Abcam, ab236505), β-Actin (Abcam, ab8226).
Cell proliferation assay
We seeded cells in 96-well plates, incubated them at 37 ℃ for 24 h, then transfected them with plasmids. Using the cell counting kit-8 (CCK-8; APExBIO, Houston, TX, USA), we performed an assay to determine cell proliferation. The CCK-8 assays were conducted at 24, 48, and 72 h. We used a microplate reader to measure the degree of yellow in the medium at an absorbance of 450 nm following the addition of CCK-8 and incubation for 2 h at 37 ℃ in a humidified CO2 incubator (Bio-Rad, Hercules, CA, USA).
Cell apoptosis analyses
An apoptosis detection Kit (KGA107, KeyGen) featuring FITC Annexin V and propidium iodide (PI) was used in this study. In 6-well plates, cells were cultivated before trypsinization. Cells were resuspended in 500 µL of binding buffer after being washed twice with PBS. A549/DDP cells were then stained for 15 min with FITC Annexin V and PI. Samples were run on a Beckman Coulter FC500 (Beckman Coulter, Brea, CA, USA). Positive Annexin V staining indicated apoptotic cells.
Migration and invasion assays
Cell migration was assessed using transwell chambers with pores of 8 µm (Corning-Costar, Corning, NY, USA). After 12 h of transfection, A549/DDP cells were digested with trypsin and suspended in appropriate amounts of serum-free 1640 medium. We then seeded 1×105 cells in 1640 basal medium in the upper chamber and in RPMI 1640 medium with 15% fetal calf serum in the lower chamber. Cells that did not migrate were removed from the transwell surface after 48 h with a cotton swab. For 20 min, we fixed the transwell membranes in paraformaldehyde at 4%, dried them, and stained them with crystal violet (0.1%, Beyotime) for 30 min. The number of cells that had passed through the cell membrane to the lower surface was determined by optical microscopy and counted using ImageJ (version 4.0.4; National Institutes of Health, Bethesda, MD, USA). An invasion assay was performed in the same manner as a cell migration assay, however, a diluted layer of Matrigel (Biozellen, Ord, NE, USA) was precoated on the membrane filter prior to the assay.
Wound healing migration assay
The 6-well plates were seeded with 5×105 cells per well 12 h prior to wound cutting. Cell monolayers were scratched in a straight line using a 200 µL pipette tip. With phase contrast microscopy, wounds were photographed immediately after wound formation and 24 h later. All optical images were analyzed using ImageJ software.
Quantification of the m6A modification
Similar to the experiment above, RNA was extracted and detected. The m6A levels of tumor/normal lung tissues and DDP-sensitive or -resistant A549 cells were measured using the EpiQuik m6A RNA Methylation Quantification Kit (P-9005-96; Epigentek, Farmingdale, NY, USA) following the manufacturer’s protocol. Assay cells were bound with 200 ng (1–8 µL) of RNA samples. After that, the capture antibody solution and detection antibody solution were added separately to the assay wells at a diluted concentration that was suitable for each. We measured each sample’s absorbance at 450 nm, using a standard curve to quantify RNA m6A.
m6A RNA immunoprecipitation
According to previously published protocols, the immunoprecipitation of m6A RNA (m6A RIP) was performed. The primers for methylated RNA immunoprecipitation (meRIP) analysis were designed based on the high-confidence fragments. For the analysis of m6A modifications, the Sangon Biotech Immunoprecipitation Kit was used (C600689; Sangon Biotech, Shanghai, China) in cisplatin‐resistant and cisplatin‐sensitive lung cancer patients’ tissues plus YTHDC2 overexpressing A549/DDP cells and common A549/DDP cells based on manufacturer’s recommendations. Isolated RNA was fragmented, and validated with 1.5% agarose gel electrophoresis, and RNA samples were then immunoprecipitated with anti-m6A or anti-mouse IgG (for control experiments) antibody-coated magnetic beads. The m6A enrichment was determined by qPCR analysis. The relative m6A level in Id3 was calculated by the m6A levels (m6A IP) normalized using the expression of itself (Input).
Bioinformatics prediction tools
The tools used for bioinformatics prediction included the following: POSTAR3 (CLIPdb) (http://111.198.139.65/RBP.html), RMVar (https://rmvar.renlab.org/) and R programming language.
Statistical analysis
Experiments were repeated 3 times with biological replicates. All statistical analyses were performed using GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA). Comparisons between 2 groups were analyzed using Student’s t-tests or analysis of variance (ANOVA). Statistical significances were considered when P values were less than 0.05. As a means of determining significance, P<0.05 was highlighted using 1 asterisk, whereas P<0.01 and P<0.001 were highlighted using 2 asterisks and 3 asterisks, separately.
Results
Id3 may be modified by m6A methylation of YTHDC2
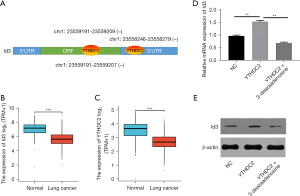
The RMVar database predicted the m6A modification of Id3, and the results showed the presence of multiple m6A modification sites in the coding sequence (CDS) region and 3’ untranslated region (3’-UTR) of Id3. The CLIPdb database found that m6A reader protein YTHDC2 can bind to Id3 (Figure 1A). The 1410 public TCGA and GTEx lung cancer cases showed that in lung cancer tissues, the expression of YTHDC2 was lower than in adjacent tissues, in accordance with Id3 (Figure 1B,1C). The above results suggested that Id3 may be modified by m6A methylation of YTHDC2, and our experimental observations confirmed this conjecture. The qPCR and WB results showed that Id3 expression was significantly up-regulated after overexpression of YTHDC2 in the cisplatin-resistant lung cancer A549/DDP cells, whereas the methylation inhibitor 3-deazaadenosine abolished the regulatory effect of YTHDC2 on Id3 (Figure 1D,1E).
YTHDC2 expression is reduced in cisplatin resistance
We used the A549/DDP cells and A549 cells to explore cisplatin-resistance in NSCLC. Our results showed that the levels of YTHDC2 mRNA was lower expressed in A549/DDP cells than in A549 cells (Figure 2A).
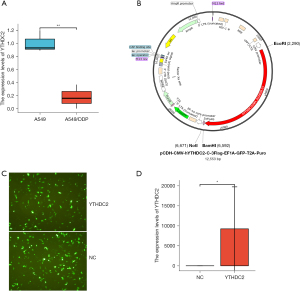
Construction and transfection of YTHDC2 vector
To validate the biological effects of YTHDC2 mimics, we constructed lentiviral vector to overexpress YTHDC2 (Figure 2B). First, we qualitatively evaluated the transfection rates of the different vectors using fluorescent microscopy and found no significant differences among them (Figure 2C). qPCR analysis confirmed that the transfection was effective (Figure 2D).
YTHDC2 overexpression suppressed the proliferation, invasion, and migration and A549/DDP cells and promoted their apoptosis
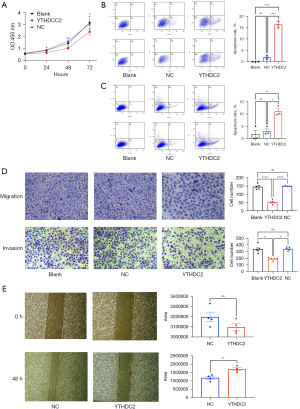
As shown in Figure 3A, the proliferation capacity of A549/DDP cells were determined with CCK-8 proliferation curves, which clearly showed that the overexpression of YTHDC2 suppressed cell proliferation. Flow cytometry assay showed a significant promotion in A549/DDP cell apoptosis after YTHDC2 overexpression at 24 and 48 h (Figure 3B,3C). Furthermore, tumor migration and invasion ability can also be used to assess tumor malignancy and influence tumor progression. The ability of tumor invasion and migration was measured using transwell assays. As well, overexpression of YTHDC2 significantly inhibited A549/DDP cells migration and invasion (Figure 3D). The above experimental results were further verified by performing wound healing assays. In Figure 3E, A549/DDP cells with overexpressed YTHDC2 displayed a significant reduction in migration capacity, as expected. The above experimental results indicated that YTHDC2 overexpression suppressed the proliferation, invasion, and migration of A549/DDP cells and promoted their apoptosis.
YTHDC2 positively regulates Id3 in an m6A-dependent manner
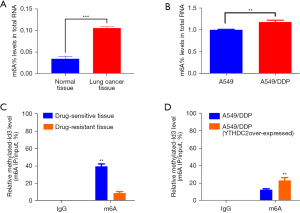
We affirmed m6A expression profiles between lung cancer and paracancerous tissues, plus between A549 and A549/DDP cells by RNA m6A colorimetry, which confirmed that m6A levels were higher in lung cancer tissues compared with normal tissues, higher in A549/DDP cells compared with A549 cells (Figure 4A,4B). The data suggested that m6A modification could contribute to cisplatin resistance in lung cancer. To determine whether YTHDC2 regulates Id3 m6A levels, immunoprecipitation of the methylated RNA was performed. Cisplatin-resistant lung cancer tissues had a lower relative methylated-Id3 level compared to cisplatin-sensitive tissues; YTHDC2 overexpressing resistant cells A549/DDP had a higher relative methylated-Id3 level compared to normal resistant cells A549/DDP (Figure 4C,4D). Based on these results, Id3’s m6A modification could be transferred in large extent by YTHDC2. In summary, YTHDC2 inhibits NSCLC resistance to cisplatin by upregulating Id3 through inhibition of m6A methylation.
Discussion
It has long been known that the development of NSCLC is a complex, multi-stage process involving the progressive adoption of genetic and epigenetic alterations, leading to unlimited growth and proliferation of tumor cells. Hence, understanding the molecular mechanisms behind NSCLC is crucial, especially in the pathogenesis of chemotherapy resistance. A number of studies have implicated m6A modification in diseases such as acute myeloid leukemia (14), hepatocellular carcinoma (15), type 2 diabetes (16), rheumatoid arthritis (17), and pulmonary hypertension (18). Recently, further studies have reported on m6A modification related protein’s biological function in lung cancer. For example, through m6A modification, METTL3 (m6A writer) promoted Bcl2 translation, increasing viability and enhancing migration in NSCLC cells (19). ALKBH5 (m6A eraser) is another m6A demethylase that promotes NSCLC progression as a result of regulating the stability of TIMP3 (20). In addition, Wang et al. demonstrated that NELL2 expression is induced by m6A demethylase FTO (m6A eraser) by inhibiting E2F1 m6A modification, leading to NSCLC metastasis (21). However, m6A binding proteins (m6A readers) remain unexplored in NSCLC.
Although YTHDC2, an m6A reader protein, has been linked to multiple cancers, its role in NSCLC is unclear. Several studies have demonstrated that the expression of YTHDC2 was significantly reduced in different cell lines of NSCLC (22). Our study found that YTHDC2 was deregulated in drug-resistant cells. However, some studies have found that YTHDC2 has high levels of expression in various cancers, including gastric cancer (23), nasopharyngeal carcinoma (12), and colon cancer (24). In future studies, we plan to investigate this mechanism further.
Uncertainty exists regarding whether cisplatin resistance in NSCLC is regulated by functional proteins involved in m6A modification. Herein, based on our functional experiments, we discovered that YTHDC2 was able to inhibit lung cancer cisplatin-resistance cells proliferation and migration, and promoted their apoptosis through m6A-dependent increases in Id3, which ultimately inhibited cisplatin resistance in lung cancer. Similarly, as Zhao et al. discovered, the expression level of YTHDC2 was positively associated with HNSCC prognoses, indicating YTHDC2 might play a HNSCC suppressor role (25). In line with our findings, Sun et al. (26) found that low expression of YTHDC2 was significantly associated with poor differentiation, lymph node metastasis, tumor size and stage by immunohistochemistry (IHC) performed in 96 NSCLC tissues and 31 adjacent normal tissues. However, a study has shown that YTHDC2 promotes colon cancer metastasis via enhancing HIF-1α mRNA translation (24). The results from these studies might seem contradictory, suggesting that as tumor types vary, YTHDC2-mediated tumor progression may act differently, which should be further investigated. Furthermore, YTHDC2 seems to affect mRNA stability in some studies. In this study, m6A-IP-PCR analysis identified YTHDC2 specifically targeted Id3, hence, we hypothesized that YTHDC2 would remove the methylation on Id3 at the transcript level. This suggested that YTHDC2 exerted a catalytic effect on Id3 protein via uninstalling the methylation and improving Id3 mRNA transcript stability. The biological roles of Id3 have been illustrated in our previous study (27,28). Yuan et al. found that YTHDC2 enhances the translation of YAP and activates oncogenic YAP signaling through its binding to the m6A coding sequence on YAP mRNA 5’-UTR (23). Our observations revealed that YTHDC2 is a new tumor suppressor in NSCLC resistant to cisplatin.
Conclusions
We have demonstrated that YTHDC2 relies on m6A modification to regulate Id3 action and ultimately inhibit cisplatin resistance in NSCLC. Hopefully, this will shed light on next-generation therapeutic strategies of NSCLC, and more studies are required in order to reveal the particular m6A regulatory mechanism.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82002233).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-187/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-187/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-187/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-187/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All participants signed an informed consent form, and this study was approved by Ethics Committee of Jinling Hospital (No. 2020DZGZRZX-096). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer 2003;3:733-44. [Crossref] [PubMed]
- Chen FF, Lv X, Zhao QF, et al. Inhibitor of DNA binding 3 reverses cisplatin resistance in human lung adenocarcinoma cells by regulating the PI3K/Akt pathway. Oncol Lett 2018;16:1634-40. [Crossref] [PubMed]
- Cantara WA, Crain PF, Rozenski J, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 2011;39:D195-201. [Crossref] [PubMed]
- Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 2014;15:293-306. [Crossref] [PubMed]
- Ye X, Wang LP, Han C, et al. Increased m(6)A modification of lncRNA DBH-AS1 suppresses pancreatic cancer growth and gemcitabine resistance via the miR-3163/USP44 axis. Ann Transl Med 2022;10:304. [Crossref] [PubMed]
- Chen H, Xiang Y, Yin Y, et al. The m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting ATG5 in seminoma. Transl Androl Urol 2021;10:1711-22. [Crossref] [PubMed]
- Liu S, Li Q, Li G, et al. The mechanism of m(6)A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis 2020;11:969. [Crossref] [PubMed]
- Shi H, Zhao J, Han L, et al. Retrospective study of gene signatures and prognostic value of m6A regulatory factor in non-small cell lung cancer using TCGA database and the verification of FTO. Aging (Albany NY) 2020;12:17022-37. [Crossref] [PubMed]
- Tsuchiya K, Yoshimura K, Inoue Y, et al. YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-small-cell lung cancer. Oncoimmunology 2021;10:1962656. [Crossref] [PubMed]
- Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res 2018;28:507-17. [Crossref] [PubMed]
- He JJ, Li Z, Rong ZX, et al. m(6)A Reader YTHDC2 Promotes Radiotherapy Resistance of Nasopharyngeal Carcinoma via Activating IGF1R/AKT/S6 Signaling Axis. Front Oncol 2020;10:1166. [Crossref] [PubMed]
- Zhou X, Han J, Zhen X, et al. Analysis of Genetic Alteration Signatures and Prognostic Values of m6A Regulatory Genes in Head and Neck Squamous Cell Carcinoma. Front Oncol 2020;10:718. [Crossref] [PubMed]
- Weng H, Huang F, Yu Z, et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell 2022;40:1566-1582.e10. [Crossref] [PubMed]
- Yang Y, Cai J, Yang X, et al. Dysregulated m6A modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol Ther 2022;30:2342-53. [Crossref] [PubMed]
- Jiang H, Yao Q, An Y, et al. Baicalin suppresses the progression of Type 2 diabetes-induced liver tumor through regulating METTL3/m(6)A/HKDC1 axis and downstream p-JAK2/STAT1/clevaged Capase3 pathway. Phytomedicine 2022;94:153823. [Crossref] [PubMed]
- Yao F, Xu C, Gao Y, et al. Expression and clinical significance of the m6A reader YTHDF2 in peripheral blood mononuclear cells from rheumatoid arthritis patients. J Immunotoxicol 2022;19:53-60. [Crossref] [PubMed]
- Liu P, Zhang A, Ding Z, et al. m(6)A Modification-mediated GRAP Regulates Vascular Remodeling in Hypoxic Pulmonary Hypertension. Am J Respir Cell Mol Biol 2022;67:574-88. [Crossref] [PubMed]
- Zhang Y, Liu S, Zhao T, et al. METTL3-mediated m6A modification of Bcl-2 mRNA promotes non-small cell lung cancer progression. Oncol Rep 2021;46:163. [Crossref] [PubMed]
- Zhu Z, Qian Q, Zhao X, et al. N(6)-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene 2020;731:144348. [Crossref] [PubMed]
- Wang Y, Li M, Zhang L, et al. m6A demethylase FTO induces NELL2 expression by inhibiting E2F1 m6A modification leading to metastasis of non-small cell lung cancer. Mol Ther Oncolytics 2021;21:367-76. [Crossref] [PubMed]
- Zhuang Z, Chen L, Mao Y, et al. Diagnostic, progressive and prognostic performance of m(6)A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci 2020;16:1785-97. [Crossref] [PubMed]
- Yuan W, Chen S, Li B, et al. The N6-methyladenosine reader protein YTHDC2 promotes gastric cancer progression via enhancing YAP mRNA translation. Transl Oncol 2022;16:101308. [Crossref] [PubMed]
- Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett 2016;376:34-42. [Crossref] [PubMed]
- Zhao X, Cui L. Development and validation of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am J Cancer Res 2019;9:2156-69. [PubMed]
- Sun S, Han Q, Liang M, et al. Downregulation of m(6) A reader YTHDC2 promotes tumor progression and predicts poor prognosis in non-small cell lung cancer. Thorac Cancer 2020;11:3269-79. [Crossref] [PubMed]
- Chen F, Zhao Q, Wang S, et al. Upregulation of Id3 inhibits cell proliferation and induces apoptosis in A549/DDP human lung cancer cells in vitro. Mol Med Rep 2016;14:313-8. [Crossref] [PubMed]
- Li XJ, Zhu CD, Yu W, et al. Overexpression of Id3 induces apoptosis of A549 human lung adenocarcinoma cells. Cell Prolif 2012;45:1-8. [Crossref] [PubMed]
(English Language Editor: J. Jones)


