Elucidation of radiation-resistant clones by a serial study of intratumor heterogeneity before and after stereotactic radiotherapy in lung cancer
Introduction
In recent years, stereotactic radiotherapy (SRT) has been effectively used to treat lung cancer, and it is steadily gaining acceptance as an alternative therapy to surgery for patients with physically inoperable conditions or those who refuse surgery (1-3). Meanwhile, there have also been sporadic reports of local recurrence and salvage surgery for local recurrence after SRT. Thus, the mechanism underlying tumor eradication by SRT warrants further investigation. In recent years, the detection of changes in mutation profiles after cancer treatment has attracted attention as a novel method for elucidating treatment efficacy (4). We encountered a patient who underwent surgery for local lung cancer recurrence after SRT. We investigated the therapeutic efficacy of SRT as a lung cancer treatment by using molecular genetic testing to compare the lung cancer mutation profiles before and after SRT.
Case presentation
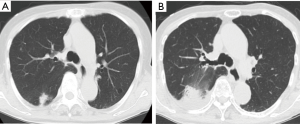
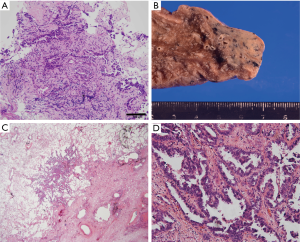
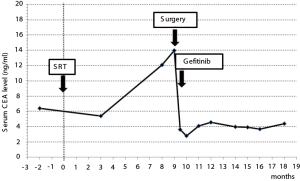
An 85-year-old woman visited our outpatient clinic for further examination of a right pulmonary nodule. Radiologic and bronchoscopic examinations revealed that the tumor was a cT1aN0M0 adenocarcinoma (Figure 1A). The patient refused surgery and chose to undergo SRT at a total radiation dose of 50 Gy in four fractions. Nine months after SRT, there appeared an area of consolidation that extended into the irradiated field (Figure 1B), and minimal pleural effusion also appeared in the right thorax. Radiologically, it was challenging to determine if the consolidated area was local tumor recurrence or prolonged inflammation due to the radiation (Figure 1B). However, because carcinoembryonic antigen (CEA) levels were persistently elevated, we suspected tumor recurrence. When we informed the patient of the suspected diagnosis, she requested surgery; thus, a right lower lobectomy and lymph node dissections were performed (Figure 2A,B). During surgery, cytology revealed malignant pleural effusion, so the operation was considered “incomplete salvage surgery”. Histopathologically, most of the resected tumor had been replaced by elastofibrosis, but the lepidic growth pattern of the adenocarcinoma was present at the periphery of the elastic lesion (Figure 2C,D). The tumor cells had invaded the visceral pleura, but no lymph node metastases were found pathologically. After surgery, serum CEA levels immediately declined to within the normal range (Figure 3). The patient was prescribed gefitinib, an epidermal growth factor receptor (EGFR)-targeted drug, and has been followed up at our outpatient clinic without any sign of recurrence to date.
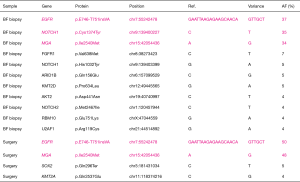
Using a next-generation sequencer, we compared mutations in the specimens biopsied by bronchoscopic examination with those in the lung tumor surgical specimens. In both samples, cancer cells were collected from formalin-fixed paraffin-embedded tissues using laser capture microdissection. Tumor purity was about 80–90%. The total amount of DNA used for next-generation sequencing analysis was 1.62 ng of the biopsy specimens and 3.76 ng of the surgical specimens (Table S1). An assay system that could analyze the entire exon region of the 53 lung cancerrelated genes (68,046 amino acids) was developed in house, and the samples were subjected to next-generation sequencing (5-7). This assay system was used to identify mutations in the 53 significantly mutated genes, which were identified from 408 patients with lung cancer by total exon analysis (8,9). In our study, mutations with an allele fraction ≥20% were deemed significant. Upon comparing mutations in the biopsy and surgical specimens, the same mutations in EGFR and MGA were detected, and interestingly, the positions and patterns of the base-pair substitutions for those mutations completely coincided (Figure 4). After SRT, the number of significant mutations decreased from three to two, and the number of insignificant mutations also decreased from eight to two (Figure 4).
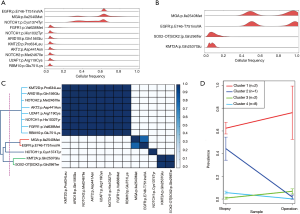
To determine whether radiotherapy changed the tumor cell composition, we performed PyClone analysis, which estimates the fraction of cancer cells harboring a mutation (Figure 5A,B) (10). Using pairwise correlation of mutations with cellular prevalence, the identified mutations were classified into four distinct clusters (Figure 5C). Of note, the cellular prevalence of clones with NOTCH1 mutations (p.Cys1374Tyr) was decreased after therapy (Figure 5D, cluster #2). In contrast, the cellular prevalence of tumor clones with EGFR and MGA mutations slightly increased after radiotherapy, indicating that these tumor clones remained after radiation therapy (Figure 5D, cluster #1). Together, these results suggest that radiosensitive or radioresistant tumor clones can coexist in a tumor (e.g., socalled intratumor heterogeneity); this heterogeneity may affect the response and resistance to radiotherapy.
This study was approved by the Institutional Ethics Board of Yamanashi Central Hospital, and the patient provided written informed consent to participate.
Discussion
In this study, we used genetic testing to investigate the therapeutic efficacy of SRT for lung cancer by comparing mutation profiles between specimens obtained before and after treatment. Although local recurrence indicated cancer progression, the number of mutations decreased, demonstrating the therapeutic efficacy of SRT. The patient, who had stage IA adenocarcinoma at the time of her initial examination, refused surgery and underwent SRT. A number of phase 2 trials demonstrated promising results from SRT, and local control rates are reportedly 80–95%, similar to surgical series (1-3). Particularly in patients ≥80 years of age, similar to the patient in our case study, definitive SRT for early-stage non-small-cell lung cancer is efficacious and safe (11,12). Two independent, randomized phase 3 trials that assessed SRT in patients with operable stage I non-small cell lung cancer closed early due to slow accrual; however, it was demonstrated that SRT could be an option for treating operable stage I non-small cell lung cancer (13). Additional randomized studies comparing SRT with surgery in patients with operable cancer are warranted.
In this case, we originally intended to perform salvage surgery; however, because pleural dissemination was detected, we were unable to perform complete curative surgery. In current clinical practice, radiological and pathological difficulties in differentiating between inflammation and local recurrence after SRT are problematic. In our case study, because the tumor was predominantly composed of elastic tissue, a differential diagnosis might have been difficult to make even if a biopsy had been performed (14-16). Therefore, the development of cancer-specific diagnostic techniques, such as genetic testing or use of genomic biomarkers, is needed.
In our study, EGFR and MGA mutations were identical in tumor specimens collected before and after SRT, suggesting that they belong to the same clone. Because of intratumor heterogeneity, tumors intrinsically contain various mutations (17,18). After exposure to radiation, it can be presumed that cancer cells with radiation-sensitive mutations are eradicated, and cancer cells with radiation-resistant mutations survive.
PyClone is a Bayesian clustering method for grouping sets of deeply sequenced somatic mutations into putative clonal clusters, while estimating their cellular prevalence and accounting for allelic imbalances introduced by segmental copy number changes and normal cell contamination (10). Thus, PyClone infers accurate clustering of mutations that co-occur in individual cells (10). In our study, PyClone analysis suggested that cells harboring NOTCH1 mutations were sensitive to radiation, whereas those harboring EGFR and MGA mutations were resistant to radiation. Recent studies suggest that activation of the NOTCH signaling pathway is related to radiation resistance in tumor cells (19-21). OncodriveROLE, a machine learning-based approach, indicated that the NOTCH1 p.Cys1374Tyr mutation, as detected in our study, is a “loss-of-function” mutation (22,23). Thus, in our case, loss-of-function in the NOTCH1 gene might have led to radiation sensitivity, resulting in the ability of cells harboring those mutations to be eradicated by radiotherapy.
We presume that this radiation-induced mutation selection resulted in decreased intratumor heterogeneity and a decreased number of detected mutations. Chemotherapy-induced changes in cancer mutation profiles have been reported, which can alter responsiveness to therapies (24-28). Here, we report that radiotherapy may also change lung cancer mutation profiles. However, because the broad clinical utility of our results has yet to be fully demonstrated, additional studies on similar cases treated with SRT are needed to validate our findings, as well as to elucidate the genomic characteristics of radiation sensitivity and resistance. In addition, the development of new therapeutic strategies to eradicate patterns of mutation in cancer, such as a combination of molecular-targeted therapy and SRT, is needed.
In conclusion, genetically identical mutations were detected in tumor specimens collected before and after SRT, and those specimens were confirmed to arise from the same clone. Meanwhile, because the number of mutations decreased after SRT, we estimated that SRT induced mutation selection. PyClone analysis showed that the therapeutic responsiveness of cancer to radiation varied among cells with different mutations, even in a single tumor. This condition is referred to as “heterogeneity in therapeutic responsiveness”, and may be responsible for the current difficulties in treating lung cancer.

Full table
Acknowledgements
The authors greatly appreciate Takuro Uchida, Hidetoshi Shigetomo, Yumi Kubota, and Ritsuko Yokouchi for their helpful scientific discussions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Informed Consent: Written informed consent was obtained from the patient for publication of this manuscript and all accompanying images.
References
- Nagata Y, Takayama K, Matsuo Y, et al. Clinical outcomes of a phase I/II study of 48 Gy of stereotactic body radiotherapy in 4 fractions for primary lung cancer using a stereotactic body frame. Int J Radiat Oncol Biol Phys 2005;63:1427-31. [Crossref] [PubMed]
- Onishi H, Shirato H, Nagata Y, et al. Stereotactic body radiotherapy (SBRT) for operable stage I non-small-cell lung cancer: can SBRT be comparable to surgery? Int J Radiat Oncol Biol Phys 2011;81:1352-8. [Crossref] [PubMed]
- Timmerman R, Paulus R, Galvin J, et al. Stereotactic body radiation therapy for inoperable early stage lung cancer. JAMA 2010;303:1070-6. [Crossref] [PubMed]
- Lambrechts S, Smeets D, Moisse M, et al. Genetic heterogeneity after first-line chemotherapy in high-grade serous ovarian cancer. Eur J Cancer 2016;53:51-64. [Crossref] [PubMed]
- Goto T, Hirotsu Y, Oyama T, et al. Analysis of tumor-derived DNA in plasma and bone marrow fluid in lung cancer patients. Med Oncol 2016;33:29. [Crossref] [PubMed]
- Goto T, Hirotsu Y, Mochizuki H, et al. Stepwise addition of genetic changes correlated with histological change from "well-differentiated" to "sarcomatoid" phenotypes: a case report. BMC Cancer 2017;17:65. [Crossref] [PubMed]
- Goto T, Hirotsu Y, Mochizuki H, et al. Mutational analysis of multiple lung cancers: Discrimination between primary and metastatic lung cancers by genomic profile. Oncotarget 2017;8:31133-43. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396-8. [Crossref] [PubMed]
- Cassidy RJ, Patel PR, Zhang X, et al. Stereotactic Body Radiotherapy for Early-stage Non-small-cell Lung Cancer in Patients 80 Years and Older: A Multi-center Analysis. Clin Lung Cancer 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Giuliani M, Hope A, Guckenberger M, et al. Stereotactic Body Radiation Therapy in Octo- and Nonagenarians for the Treatment of Early-Stage Lung Cancer. Int J Radiat Oncol Biol Phys 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Chang JY, Senan S, Paul MA, et al. Stereotactic ablative radiotherapy versus lobectomy for operable stage I non-small-cell lung cancer: a pooled analysis of two randomised trials. Lancet Oncol 2015;16:630-7. [Crossref] [PubMed]
- Senthi S, Lagerwaard FJ, Haasbeek CJ, et al. Patterns of disease recurrence after stereotactic ablative radiotherapy for early stage non-small-cell lung cancer: a retrospective analysis. Lancet Oncol 2012;13:802-9. [Crossref] [PubMed]
- van den Berg LL, Klinkenberg TJ, Groen HJ, et al. Patterns of Recurrence and Survival after Surgery or Stereotactic Radiotherapy for Early Stage NSCLC. J Thorac Oncol 2015;10:826-31. [Crossref] [PubMed]
- Huang K, Dahele M, Senan S, et al. Radiographic changes after lung stereotactic ablative radiotherapy (SABR)--can we distinguish recurrence from fibrosis? A systematic review of the literature. Radiother Oncol 2012;102:335-42. [Crossref] [PubMed]
- Welch DR. Tumor Heterogeneity--A 'Contemporary Concept' Founded on Historical Insights and Predictions. Cancer Res 2016;76:4-6. [Crossref] [PubMed]
- Zellmer VR, Zhang S. Evolving concepts of tumor heterogeneity. Cell Biosci 2014;4:69. [Crossref] [PubMed]
- Shen Y, Chen H, Zhang J, et al. Increased Notch Signaling Enhances Radioresistance of Malignant Stromal Cells Induced by Glioma Stem/ Progenitor Cells. PLoS One 2015;10:e0142594. [Crossref] [PubMed]
- Yahyanejad S, King H, Iglesias VS, et al. NOTCH blockade combined with radiation therapy and temozolomide prolongs survival of orthotopic glioblastoma. Oncotarget 2016;7:41251-64. [Crossref] [PubMed]
- Yahyanejad S, Theys J, Vooijs M. Targeting Notch to overcome radiation resistance. Oncotarget 2016;7:7610-28. [Crossref] [PubMed]
- Schroeder MP, Rubio-Perez C, Tamborero D, et al. OncodriveROLE classifies cancer driver genes in loss of function and activating mode of action. Bioinformatics 2014;30:i549-55. [Crossref] [PubMed]
- Southgate L, Sukalo M, Karountzos AS, et al. Haploinsufficiency of the NOTCH1 Receptor as a Cause of Adams-Oliver Syndrome With Variable Cardiac Anomalies. Circ Cardiovasc Genet 2015;8:572-81. [Crossref] [PubMed]
- Morrissy AS, Garzia L, Shih DJ, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016;529:351-7. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Song HN, Jung KS, Yoo KH, et al. Acquired C797S Mutation upon Treatment with a T790M-Specific Third-Generation EGFR Inhibitor (HM61713) in Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:e45-7. [Crossref] [PubMed]
- Tan SH, Sapari NS, Miao H, et al. High-Throughput Mutation Profiling Changes before and 3 Weeks after Chemotherapy in Newly Diagnosed Breast Cancer Patients. PLoS One 2015;10:e0142466. [Crossref] [PubMed]
- Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40. [PubMed]


