Research status and funding trends of lung cancer biomarkers
According to 2008 World cancer report issued by International Agency for research on cancer (IARC) that contained information of cancer morbidity and mortality, there were estimated to be 1,608,055 cases of lung cancer worldwide in 2008, nearly 16.5% of all new cancer cases; 1,376,579 deaths, representing 22.5% of all cancer, as a top leading cause of cancer deaths (1). In the report of third leading cause of death nationwide sample survey by China’s Ministry of Health, lung cancer was the leading cause of death in all the cancer registries, with the mortality of 30.83/100,000; estimated number of new cases of lung cancer nationwide in 2005 was 536,407. Lung cancer mortality markedly elevated, accounting to 22.7% of total cancer deaths, as a top cause of cancer deaths (2). According to World Health Organization’s (WHO) global estimates, the registered lung cancer mortality rate increased by 464.84% in past three decades. Annual lung cancer deaths in China will reach about one million by 2005 (3).
Although much progress has been recently made toward the methods and strategies to lung cancer treatment, the survival rate of lung cancer patients has not been significantly improved, but challenges remain for solving this problem (4). Therefore, it is essential to develop biomarkers for early diagnosis of lung cancer, prediction of tumor metastasis, and evaluation of treatment efficiency, as well as to translate highly promising findings in basic science research to clinical application (5,6). Since substantial progress in genomics, proteomics and transcriptomics has been achieved, those new technologies greatly affect cancer-related research. As a result, the identification of a variety of biomarkers for lung cancer becomes more convenient, accelerating the development of novel tumor biomarkers and screening methods for therapeutic targets (7-11). In this review, author discussed the status of lung cancer research on biomarkers and the funding trends worldwide in this research field.
Biomarkers for early diagnosis of lung cancer
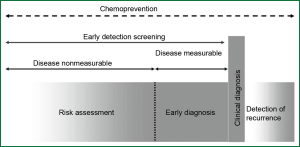
Lung cancer may be caused by a long-term respiratory damage and repair repeatedly. It is a rather lengthy period for lung cancer patients from onset of the disease to tumor development until clinical symptoms appear and diagnosis is made. Therefore, early diagnosis and risk assessment performed in the days before clinical symptoms appear, is one of the most effective ways to improve survival rate of lung cancer patients (12) (Figure 1). During the multi-stage carcinogenesis of lung cancer, there used to be a serious of mutations of oncogenes and tumor suppressor genes as well as aberrant epigenetic regulation of gene expression, which led to subsequent abnormal gene transcription and translation. Many factors involved in those processes could serve as biomarkers for early diagnosis or early screening tools for lung cancer (13,14).
Gene mutation and nucleotide polymorphism-related tumor biomarkers
Since gene mutations, single nucleotide polymorphisms (SNP), and abnormal gene copy number (copy number variations, CNVs) are closely correlated with tumorigenesis and tumor development, those molecular abnormalities can be used as biomarkers for early diagnosis of lung cancer. For example, decreased expression of HYAL2, FHIT and SFTP is often found in tumor and saliva specimens from patients with non small cell lung cancer (NSCLC). Li et al. (15) reported that with technique of fluorescence in situ hybridization (FISH), genetic aberrations in the genes HYAL2, FHIT, and SFTPC were investigated in paired tumors and saliva samples. They found that genetic deletions of HYAL2 and FHIT in saliva can be used as early diagnostic markers for NSCLC patients with smoking history. DNA repair gene XRCC4 plays an important role in the non-homologous end-joining system within tumor cells. In Hsu’s study (16), 7 candidate SNPs in XRCC4 gene were examined in samples from lung cancer patients and healthy controls in the population of central Taiwan. Among those SNPs, XRCC4 G-1394T (rs6869366) was identified as a high risk factor in patients with smoking history, as compared to other subjects (OR =2.31, 95% CI =1.43-3.72), implicating that the G allele of XRCC4 G-1394T may serve as a marker for early diagnosis and a target for lung cancer prevention. In Rosenberger’s report (17), his results suggest that polymorphism of GPX1 C-599T (rs1050450) and EPHX C-337T (rs1051740) is associated with the susceptibility to patients with early stage lung cancer, younger than 51 years at diagnosis. Massion et al. (18) reported, that genomic gains in specific loci have been found by FISH assay in bronchial biopsy specimens from lung cancer patients, including increased gene copy number of TP63 (3q28), MYC (8q245p15.2), and centromeric regions for chromosome 3 (CEP3) and 6 (CEP6). Combination of these 4 specific probes offered a sensitivity of 82% for lung cancer prediction and specificity of 58%. Those specific cytogenetic alternations can be used as biomarkers for the early diagnisis of lung cancer and have value in assessing lung cancer risk.
Epigenetic modulation and tumor biomarkers
An increasing body of evidence (19-21) clearly indicate that aberrant gene expression in the absent of changes in genome nucleotide sequence, such as DNA methylation of genes, and dysregulated microRNAs, can be used as biomarkers for early diagnosis of lung cancer. Lokk et al. (22) performed a DNA methylation profiling using microarray that covers the promoter regions of more than 14,500 genes. The results showed hypermethylation of 496 CpGs in 379 genes and hypomethylation of 373 CpGs in 335 genes in stage 1 NSCLC. Among those identified genes with altered methylation, a lot of the loci with altered methylation can be considered as mark candidates for the molecular screening of early stage NSCLC. According to Geng’s report (23), methylation analysis of varieties of genes involved in NSCLC revealed that detection with an optimized 5-gene panel (NEUROG2, NID2, RASSF1A, APC and HOXC9) can improve the diagnostic potential for stage 1 NSCLC and achieved a sensitivity of 91.26% and specificity 84.62%. Zhao et al. (24) analyzed the methylation status of 13 genes in specimens from stage 1 NSCLC patients and the normal controls. They identified 7 hypermethylated genes and 5 hypomethylated genes in their study. Richards et al. found (25) that the promoter of transcription factor TCF21 promoter has been hypermethylated in NSCLC. 81% of NSCLC samples showed TCF21 promoter hypermethylation, which can be used as a candidate marker for early-stage NSCLC.
Recent studies (26,27) found that miRNAs are involved in the pathogenesis of lung cancer. Xing et al. (28) analyzed data from miRNA expression profiling regarding squamous lung cell carcinoma in sputum and found a panel of microRNA markers (combination of miR-205, miR-210 and miR-708) suitable for the early diagnosis of lung squamous cell carcinoma patients. By logistic regression analysis, Tang et al. (29) identified 3 plasma miRNAs including miR-21, miR-145 and miR-155 as noninvasive biomarkers for early detection of lung cancer.
Gene transcripts and tumor biomarkers
Oncogene activation and tumor suppressor gene inactivation in lung cancer are essential to tumorigenesis of lung cancer. Aberrant expression products during tumorigenesis can be used as candidate biomarkers for the early diagnosis of lung cancer, such as CEA (carcino-embryonic antigen), a well-known tumor marker. Nosotti et al. (30) evaluated the correlation of CEA mRNA level with micrometastases in lymph nodes and 5-year survival rate in stage 1 NSCLC, implicating that CEA mRNA expression levels can be used as a molecular detection of early stage lung cancer. According to this finding, the expression level of CEA mRNA is suggested to be a useful molecular marker for early-stage lung cancer. Survivin and livin are two members of inhibitor of apoptosis gene family. Li et al. (31) evaluated the diagnostic role of Survivin and livin mRNA expression in the bronchial aspirates of patients with lung cancer, via analysis of receiver operating characteristic curve (ROC), indicating elevated mRNA expression of both Survivin and livin may be valuable biomarker for the early diagnosis of lung cancer. The expression profiling of mRNA with microarray is also applied in the early detection of lung cancer. In a previous report (32), an investigation on mRNA expression profiling was performed by microarray in double blind test of 442 lung adenocarcinoma samples. Clinical and molucular models of lung cancer established based on the microarray data obtained in that study have predictive value for patients with early-stage lung cancer. Lu et al. (33) applied a meta-analysis of datasets from 7 different microarray studies on differentially expressed genes in NSCLC. They identified a gene expression signature consisting of 64 genes that is helpful in predicting stage 1 lung cancer patients as well as making decision for the further invasive therapy.
Gene translation products and tumor biomarkers
Due to transcriptional and post-translational regulation, the number of protein products far exceeds the number of genes and proteins are more stable than mRNA. As proteomic technologies are rapidly growing, it makes possible to perform large-scale lung cancer screening, so that candidate biomarkers for lung cancer detection can be finally validated. Rahman et al. (34) identified a specific proteomic profile that allows an overall predictive accuracy of over 90% of normal, preinvasive, and invasive lung cancer, in analysis of several different matrix-assisted laser desorption/ionization time-of-flight mass spectrometry profiles. They found that the specific patterns of protein expression of the airway epithelium can accurately classify normal bronchial and alveolar tissues from preinvasive bronchial lesions and also from invasive lung cancer. In their studies, the mass-to-charge ration features of 9 different matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) profiles were further analyzed. Results showed the mass-to-charge ration features associated with brochial lesions might benefit the early diagnosis and risk assessment of lung cancer (35). Ostroff et al. (36) conducted a multi-center case-control study in archieved serum samples from 1,326 subjects, with a new aptamer-based proteomic technology. They identified a 12-protein panel including cadherin-1, CD30 ligand, endostatin, HSP90α, LRIG3, MIP-4, pleiotrophin, PRKCI, RGM-C, SCF-sR, sL-selectin and YES, which can efficiently discriminates NSCLC from controls, also distinguish early and late stage NSCLC. Since auto-antibodies can recognize tumor-associated antigens (aberrant cancer proteins) within human cells, tumor-specific autoantibody profiling is desirable for development of a new diagnositic assay. Khattar et al. (37,38) developed a multiplex assay for autoantibody profiling that can recognize NSCLC-associated antibodies, with potential for predicting NSCLC.
Biomarkers for prognosis or metastasis of lung cancer
Lung cancer has become one of human malignancies with highest morbidity and mortality rates in 21st century. In 2012 annual report of China Cancer Registry, 5-year survival rate for lung cancer patients is less than 16%, which is closely related to lung cancer metastasis. Therefore, it is possible to extend the survival time of lung cancer patients to a certain extent, by accuracy assessment of the prognostic factors, such as patients’ history and pathological patterns; development of predictive biomarkers for prognosis and risk or metastasis in different individuals, and molecular targeted therapy based on molecular typing of lung cancer. At present, remarkable progress has been made in diagnosis and treatment of lung cancer, including novel lung cancer-associated biomarkers identified by new technologies of genomics and proteomics as well as molecular targeted small-molecular anti-cancer drugs related to those novel biomarkers. Haruki et al., (39) reported overexpression of geminin in small lung adenocarcinoma (AC). It has been found that geminin is a small- molecular multiple functional protein located at nucleus, which plays important roles in cell proliferation, embryonic development and tumorigenesis, implicating that serum level of geminin may be a useful prognostic indicator in patients with lung AC and geminin may become a promising target for lung cancer treatment. Serum amyloid A (SAA) is known as an acute phase protein and it is also a polymorphic protein in the family of apolipoproteins. SAA has been identified and validated as a potential lung cancer biomarker by analysis of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS). In recent report, SAA level in NSCLC patients who survived ≥5 years is significantly lower than that in NSCLC patients who survived <5 years, suggesting elevated SAA may be as a non-invasive biomarker to predict the prognosis of NSCLC patients (40). Furthermore, vascular epidermal growth factor (VEGF) is closely associated with tumor growth. Jantus-Lewintre (41) found a poor clinical outcome in NSCLC patients with up-regulated VEGF and down-regulated vascular endothelial growth factor receptor 2 (VEGFR2). Whereas, Pajares et al. (42) found that an increase in VEGF and vascular endothelial growth factor receptor 1 (VEGFR1) and VEGFR2 incaticated a favorable prognosis in patients with early-stage lung squamous cell carcinoma but expression levels of those factors are not related to prognosis of patients with lung adenocarcinoma.
Part 2
Chang et al. (43,44) reported that increased level of VEGF and decreased level of thrombospondin-1 (TSP-1) correlate with poor prognosis in NSCLC patients. VEGF expression is associated with inflammatory factors, such as sialic acid and IL-6. Currently inhibitors targeting the prognostic marker, epithelial growth factor receptor (EGFR-TKls) has been widely used in cancer treatment, such as gefitinib (Iressa), and erlotinib (Tarceva). Phosphorylation of tyrosine kinases and signal transduction of the downstream effectors can be inhibited by EGFR-TKls, to which patients with EGFR mutations are sensitive. In literatures, (45) oncogenic mutation occurring in RAS family members correlate with poor prognosis. P21 of ras gene product plays a key role in cell proliferation and differentiation. Lung cancer patients with ras mutation have shorter survival times as compare to those without ras mutation. Kras mutation has been found in 20-30% NSCLC patients (46). A recent study (47) showed that MEK, a potent Ras downstream effector can be inhibited by selumetinib (AZD6244) that has been used for treatment of head and neck tumors with Kras mutation, leading to a remarkable prolonged survival in cancer patients. Recently, ROS1 rearrangement has been identified in 2% NSCLC patients (48) and clinical trial showed that crizotinib has promise for stage 1 lung cancer patients with the ROS1 rearrangement. Crizotinib is a multi-target inhibitor against ROS1, MEK and EML4-ALK and becomes a significant drug of molecular targeted therapy in NSCLC. 2011 crizotinib was approved by FDA for the treatment of patients with advanced ALK-positive NSCLC.
MiRNA is a class of single-strand small noncoding RNAs that have been recently found closely associated with tumorigenesis and cancer development. Therefore, miRNA has the potential to become novel tumor biomarkers for predicting prognosis and therapeutic efficacy. Wang et al. (49) initially reported elevated miRNA-130a in NSCLC tissue, which was identified as an independent prognostic indicator for NSCLC patients and also closely associated with lymph node metastasis, tumor staging and prognosis. According to Hu’s investigation (50), the serum levels of 11 miRNAs including miR-486 and miR-22 in lung cancer patients with a short-term survival was 5 times higher than those with a long-term survival, indicating elevated miRNAs is closed to survival of Lung cancer patients, also providing the proof that dysregulated serum miRNAs as potential biomarkers can predict development and prognosis of lung cancer.
Funding trends home and abroad in research fields of cancer biomarkers
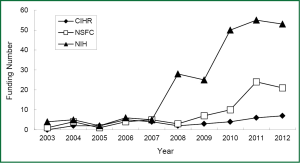
Since 2003, there is a trend in annually growing number of submitted grant application regarding lung cancer biomarkers to National Institute of Health (NIH, United States), Canadian Institutes of Health Research (CIHR) and National Natural Science Foundation of China (NSFC). For example, if you search the projects using keyword “lung cancer” and “biomarker”, you will find a rapid increase in granted projects in NIH since 2010, the number of granted projects from 4 in 2003 up to 53 in 2012. Similarly, there is the growing number of granted projects in both CIHR and NSFC, but their number is obviously less than that in NIH. Although the granted projects in NSFC are increasing from 4 in 2004 to 21 in 2012, the gap of the growth rate is still large between NSFC and NIH (Figure 2). As shown in a diagram of funding situation, nearly 10 years the domestic and international granted projects in the field of lung cancer biomarkers have gradually increased, especially significantly increased in the last 5 years. However, the growing rate of funding in this field is relatively smaller in China, possibly due to relatively fewer basic research groups in this field.
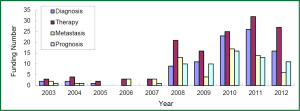
The research direction of granted projects covered early diagnosis, therapy, and predictive indicators for metastasis and prognosis of lung cancer. In growing petition for the grant related to lung cancer biomarkers received by NSFC, NIH, and CIHR since 2003, highly attention has been paid to the projects in terms of early diagnosis and molecular targeted therapy for lung cancer, which readily received more funding (Figures 3-5). Among the NIH granted research projects of lung cancer in 2012, there are 16 projects regarding biomarkers for early diagnosis, 27 projects of molecular targeted therapy, 6 projects related to metastasis and 11 projects for prognostic indicators. It is apparent that early diagnosis and molecular targeted therapy of lung cancer are currently hot spot in the field of lung cancer biomarker research.
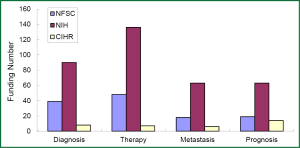
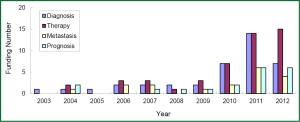
Although the growing rate of funding in NSFC is less than that that in NIH, several research groups in this field have been developed through financial support by NSFC grants, some of which were supported by key projects fund. For example, the team led by Prof. Shen Hongbing at Nanjing Medical University performed cohort studies of the identification of genetic variants associated with common malignancies and risk prediction. Those studies were really sponsored by key projects fund. By means of genome-wide association study (GWAS), significant achievements has been made by this team in the projects of the association of genetic susceptibility of gastric and lung cancer with early diagnosis and cancer treatment. Their research results have been reported in Nat Genet (51-54). Another example is new progress of the study in Prof. Huang Gang’s research team at Shanghai Jiaotong University, on serum Dickkop-1 (DKK1) as a novel biomarker for lung cancer detection (55). Research group of lung cancer for clinical translation at Cancer Hospital, Medical Center of Fudan University, led by Prof. Chen Haiquan developed a rapid and accurate method of molecular diagnosis to detect ALK-rearranged lung cancer. Clinical application of this new method will not only reduce a lot of expensive testing costs for cancer patients, but also improve opportunity of personalized molecular targeted therapy to advanced lung cancer patients (56). A genetic mapping of pathogenicity determinants in non-smoking lung cancer patients has been completed by the collaboration between Prof. Chen Haiquan’s team and Prof. Ji Hongbin in Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS). These findings could provide theoretical basis and practical guidance for personalized cancer therapy to non-smoking lung cancer patients (57). A report in Nat. Genet by Prof. Lin Dongxin at Cancer Institute and Hospital, Chinese Academy of Medical Science, revealed that a six-nucleotide deletion variant in the CASP8 promoter is associated with decreased risk of lung cancer. The identified deletion variant in the CASP8 promoter correlates with reduced susceptibility in Chinese population to multiple tumors, including lung, esophageal, gastric, colorectal, cervical and breast cancers (58).
Future prospects
Recent studies find a critical issue that most of tumor biomarkers may appear in multiple organs or tissues as well as in both disease and health status, when a single biomarker is used for early diagnosis and prognosis of lung cancer. Therefore, the detection with a single tumor biomarker is difficult to simultaneously meet the requirements of both high sensitivity and specificity. Detection with combined cancer biomarkers will be more meaningful. Since development of lung cancer is a complex, multiphase and multi-step pathological process with changes in all aspects of transcription, translation and regulation of gene expression, it is necessary to identify more specific biomarkers of lung cancer by expanding sample size and develop more reliable and accurate methods using biomarkers for diagnosis of lung cancer and prediction of the therapeutic efficacy. The progress in this aspect will greatly promote the development of personalized medicine in lung cancer. But, immense challenges still remain in the clinical translation studies of tumor biomarkers as well as the diagnosis and prognosis with combined cancer biomarkers (3).
As the funding strength increases with years, rapid progress was made in the field of lung cancer biomarkers. It is desirable to develop the biomarkers that are directly associated with tumorigenesis and development of lung cancer, via technologies of high-throughput sequencing and bioinformatics analysis. Meanwhile, to design more molecular targeted therapy with high specificity and less adverse drug reaction, based on novel tumor biomarkers is also a high priority in lung cancer research. The studies of translational medicine and personalized medicine are also strongly encouraged, because improvements in this aspect will finally convert achievements of basic research into clinical practice. All these research directions will be undoubtedly primary focus of funding.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [PubMed]
- Chen WQ, Zhang SW, Zou XN, et al. An analysis of lung cancer mortality in China, 2004 - 2005. Zhonghua Yu Fang Yi Xue Za Zhi 2010;44:378-82. [PubMed]
- She J, Yang P, Hong Q, et al. Lung cancer in China: challenges and interventions. Chest 2013;143:1117-26. [PubMed]
- Sekhon HS, Souza CA, Gomes MM. Advances in cytopathology for lung cancer: the impact and challenges of new technologies. Thorac Surg Clin 2013;23:163-78. [PubMed]
- Hirsch FR, Franklin WA, Gazdar AF, et al. Early detection of lung cancer: clinical perspectives of recent advances in biology and radiology. Clin Cancer Res 2001;7:5-22. [PubMed]
- Cho WC. Molecular diagnostics for monitoring and predicting therapeutic effect in cancer. Expert Rev Mol Diagn 2011;11:9-12. [PubMed]
- Van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature 2008;452:564-70. [PubMed]
- Cho WC. Contribution of oncoproteomics to cancer biomarker discovery. Mol Cancer 2007;6:25. [PubMed]
- Chen HY, Yu SL, Li KC, et al. Biomarkers and transcriptome profiling of lung cancer. Respirology 2012;17:620-6. [PubMed]
- Hassanein M, Callison JC, Callaway-Lane C, et al. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5:992-1006. [PubMed]
- Toraño EG, Petrus S, Fernandez AF, et al. Global DNA hypomethylation in cancer: review of validated methods and clinical significance. Clin Chem Lab Med 2012;50:1733-42. [PubMed]
- Hassanein M, Rahman JS, Chaurand P, et al. Advances in proteomic strategies toward the early detection of lung cancer. Proc Am Thorac Soc 2011;8:183-8. [PubMed]
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet 1993;9:138-41. [PubMed]
- Li G, Xiao Z, Liu J, et al. Cancer: a proteomic disease. Sci China Life Sci 2011;54:403-8. [PubMed]
- Li R, Todd NW, Qiu Q, et al. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin Cancer Res 2007;13:482-7. [PubMed]
- Hsu NY, Wang HC, Wang CH, et al. Lung cancer susceptibility and genetic polymorphism of DNA repair gene XRCC4 in Taiwan. Cancer Biomark 2009;5:159-65. [PubMed]
- Rosenberger A, Illig T, Korb K, et al. Do genetic factors protect for early onset lung cancer? A case control study before the age of 50 years. BMC Cancer 2008;8:60. [PubMed]
- Massion PP, Zou Y, Uner H, et al. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PLoS One 2009;4:e5611. [PubMed]
- Oak CH, Wilson D, Lee HJ, et al. Potential molecular approaches for the early diagnosis of lung cancer Mol Med Rep 2012;6:931-6. [PubMed]
- Pass HI, Beer DG, Joseph S, et al. Biomarkers and molecular testing for early detection, diagnosis, and therapeutic prediction of lung cancer. Thorac Surg Clin 2013;23:211-24. [PubMed]
- Balgkouranidou I, Liloglou T, Lianidou ES. Lung cancer epigenetics: emerging biomarkers. Biomark Med 2013;7:49-58. [PubMed]
- Lokk K, Vooder T, Kolde R, et al. Methylation markers of early-stage non-small cell lung cancer. PLoS One 2012;7:e39813. [PubMed]
- Geng J, Sun J, Lin Q, et al. Methylation status of NEUROG2 and NID2 improves the diagnosis of stage I NSCLC. Oncol Lett 2012;3:901-6. [PubMed]
- Zhao Y, Zhou H, Ma K, et al. Abnormal methylation of seven genes and their associations with clinical characteristics in early stage non-small cell lung cancer. Oncol Lett 2013;5:1211-8. [PubMed]
- Richards KL, Zhang B, Sun M, et al. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer 2011;117:606-17. [PubMed]
- Wu X, Piper-Hunter MG, Crawford M, et al. MicroRNAs in the pathogenesis of Lung Cancer. J Thorac Oncol 2009;4:1028-34. [PubMed]
- Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430-41. [PubMed]
- Xing L, Todd NW, Yu L, et al. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol 2010;23:1157-64. [PubMed]
- Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev 2013;22:540-8. [PubMed]
- Nosotti M, Palleschi A, Rosso L, et al. Lymph node micrometastases detected by carcinoembryonic antigen mRNA affect long-term survival and disease-free interval in early-stage lung cancer patients. Oncol Lett 2012;4:1140-4. [PubMed]
- Li J, Chen P, Li XQ, et al. Elevated levels of survivin and livin mRNA in bronchial aspirates as markers to support the diagnosis of lung cancer. Int J Cancer 2013;132:1098-104. [PubMed]
- Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma, Shedden K, Taylor JM, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [PubMed]
- Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med 2006;3:e467. [PubMed]
- Rahman SM, Shyr Y, Yildiz PB, et al. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med 2005;172:1556-62. [PubMed]
- Rahman SM, Gonzalez AL, Li M, et al. Lung cancer diagnosis from proteomic analysis of preinvasive lesions. Cancer Res 2011;71:3009-17. [PubMed]
- Ostroff RM, Bigbee WL, Franklin W, et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One 2010;5:e15003. [PubMed]
- Khattar NH, Coe-Atkinson SP, Stromberg AJ, et al. Lung cancer-associated auto-antibodies measured using seven amino acid peptides in a diagnostic blood test for lung cancer. Cancer Biol Ther 2010;10:267-72. [PubMed]
- Lam S, Boyle P, Healey GF, et al. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 2011;4:1126-34. [PubMed]
- Haruki T, Shomori K, Hamamoto Y, et al. Geminin expression in small lung adenocarcinomas: implication of prognostic significance. Lung Cancer 2011;71:356-62. [PubMed]
- Cho WC, Yip TT, Cheng WW, et al. Serum amyloid A is elevated in the serum of lung cancer patients with poor prognosis. Br J Cancer 2010;102:1731-5. [PubMed]
- Jantus-Lewintre E, Sanmartin E, Sirera R, et al. Combined VEGF-A and VEGFR-2 concentrations in plasma: diagnostic and prognostic implications in patients with advanced NSCLC. Lung Cancer 2011;74:326-31. [PubMed]
- Pajares MJ, Agorreta J, Larrayoz M, et al. Expression of tumor-derived vascular endothelial growth factor and its receptors is associated with outcome in early squamous cell carcinoma of the lung. J Clin Oncol 2012;30:1129-36. [PubMed]
- Chang A, Parikh P, Thongprasert S, et al. Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non-small cell lung cancer: subset analysis from the ISEL study. J Thorac Oncol 2006;1:847-55. [PubMed]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123-32. [PubMed]
- Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92:131-9. [PubMed]
- Bacchi CE, Ciol H, Queiroga EM, et al. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics (Sao Paulo) 2012;67:419-24. [PubMed]
- Metro G, Chiari R, Baldi A, et al. Selumetinib: a promising pharmacologic approach for KRAS-mutant advanced non-small-cell lung cancer. Future Oncol 2013;9:167-77. [PubMed]
- Rothschild SI, Gautschi O. Crizotinib in the treatment of non-small-cell lung cancer. Clin Lung Cancer 2013;14:473-80. [PubMed]
- Wang XC, Du LQ, Tian LL, et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer 2011;72:92-9. [PubMed]
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 2010;28:1721-6. [PubMed]
- Lan Q, Hsiung CA, Matsuo K, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 2012;44:1330-5. [PubMed]
- Dong J, Hu Z, Wu C, et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat Genet 2012;44:895-9. [PubMed]
- Shi Y, Hu Z, Wu C, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet 2011;43:1215-8. [PubMed]
- Hu Z, Wu C, Shi Y, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011;43:792-6. [PubMed]
- Sheng SL, Huang G, Yu B, et al. Clinical significance and prognostic value of serum Dickkopf-1 concentrations in patients with lung cancer. Clin Chem 2009;55:1656-64. [PubMed]
- Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008;14:4275-83. [PubMed]
- Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010;28:4616-20. [PubMed]
- Sun T, Gao Y, Tan W, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet 2007;39:605-13. [PubMed]


