Gene methylation in liquid biopsy and risk of recurrence in lung cancer
Belinsky and colleagues showed the potential utility for predicting tumour recurrence of an 8-gene methylation panel in plasma and sputum from resected Stage I non-small cell lung cancer (NSCLC) patients (1). In the prevention study ECOG-ACRIN5597, 1,561 patients were randomized to receive placebo or L-selenomethionine treatment daily for 4 years. The primary endpoint of the study was to evaluate the effectiveness of selenium for reducing the incidence of second primary tumours. A biomarker sub-study in 565 patients was designed to analyze the prevalence for methylation of the 8-gene signature (P16, MGMT, DAPK, RASSF1A, GATA4, GATA5, PAX5α and PAX5β) in both sputum and plasma using methylation-specific PCR (MSP). The author demonstrated that subjects with one or more genes methylated in plasma had an increased risk for local or distant recurrence compared to those patients without methylated genes. This association was not observed in sputum, in which methylation prevalence was much higher than in plasma. A significant association between recurrence independently of location (local or distant) and risk of gene methylation (1.4-fold increase in odds ratio) in sputum was found only in the placebo-treated group, but this association was not confirmed in plasma samples.
Currently, the standard treatment for stage I NSCLC is surgery. However, approximately 30% of patients with pathological stage IA/IB NSCLC still have either local recurrence or distant metastasis (2,3). The risk factors resulting in NSCLC treatment failure after surgery have not been elucidated and advancements in early diagnosis are still awaited. It is noteworthy to mention that early diagnosis and treatment of lung cancer improves either short-term or long-term outcomes. Therefore, the search for new biomarkers in this setting is particularly necessary.
One of the most surprising observations in molecular medicine was the discovery that genes’ expression was altered by epigenetic mechanisms (4). Epigenetics changes are not attributable to changes in DNA sequence, are present almost in all human cancers, are inextricably linked with genetic alterations, and individually or jointly can cause the development of a malignant phenotype. The most frequent epigenetic alteration is methylation of the gene promoter region that is generally hypermethylated in cancer. This epigenetic change negatively affects the transcription of a set of genes involved in all cellular functions and might be considered as an alternative way for the loss of gene function in cancer. When this phenomenon occurs, it is also accompanied by changes in chromatin composition around short interspersed DNA dinucleotide CpG sequences (GC-rich, CpG-rich) called CPG ISLAND, located near to the transcription initiation. These chromatin structures are characterized also by the acetylation, phosphorylation, methylation, and ubiquitylation of histone tails and all these modifications act by preventing the access to the transcriptional machinery (5,6).
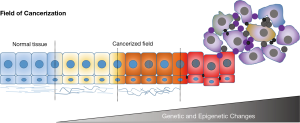
Recently the analysis of gene-promoter hypermethylation has become object for the development of new molecular strategies to provide diagnostic tools for the early detection and prognosis in cancer. In this respect, Belinsky and colleagues (7) previously proposed a sensitive DNA-based assay, based on the detection of a panel of methylated genes in the sputum through MSP, to evaluate epigenetic alterations in smokers. MSP technique allows for highly sensitive detection of locus-specific DNA methylation, using PCR amplification of bisulfite-converted DNA. In particular, a nested-based approach (nested MSP) allows to assess the methylation status of DNA deriving from few cells (8). In their earlier study, Belinsky and collaborators found that promoters’ gene methylation in exfoliated cells from the lungs could predict cancer up to 18 months prior to clinical diagnosis (7). Indeed, the analysis of DNA methylation in lung cancer cells provides an assessment of the extent of the “field cancerization”. The concept of field cancerization was first introduced in the early fifties by Slaughter et al., who analyzed the tissues adjacent to squamous cell carcinoma (9). According to this hypothesis, prior to the growth of a cancerous lesion, a normal cell lineage can acquire pro-tumorigenic ‘capabilities’ due to genetic mutations or epigenetic changes that are positively selected within the microenvironment. As result, this normal-appearing tissue can grow and produce large fields of cells that are predisposed to eventually progress to a neoplasm (10).
A recent definition of a cancerized field was “a collection of cells that have gained, some but not all, the phenotypic alterations required for malignancy”. Collectively, these changes might not be detected as morphological abnormalities and might include properties such as an increased growth rate, decreased death rate or increased immune evasion (11) (Figure 1). Therefore, biomarkers reflecting the progression of a cancerized field are likely to predict tumour development. The finding that these biomarkers can be easily detected in plasma and sputum and do correlate with the outcome of NSCLC patients represents a significant innovation in this field.
There is an increasing interest in the use of liquid biopsy for early detection of cancer (12-15). Liquid biopsy is a quite broad term that refers to the possibility to measure tumour biomarkers in patients’ derived body fluids. Although blood is the main approach for liquid biopsy, other fluids including sputum, urine, cerebrospinal liquor might offer advantages for biomarker detection, depending on the anatomic localization of the tumour (Table 1). In this regard, different studies assessed sputum as non-invasive screening method for investigating respiratory tract malignancy for decades. Saccomanno and colleagues showed that sputum cytology was able to detect premalignant changes in high-risk groups several years before a clinical diagnosis of lung cancer (16). However, the detection of morphological changes in the cytology of bronchial cells requires skills that are subjective of the pathologist performing the analysis, thus raising concerns about its reliability and reproducibility. Another limitation is that the identification of aggressive tumours where the cells look very different from normal cells is easier than the identification of low-grade, non-invasive tumours where the individual cells look normal, making this approach of little clinical utility. Therefore, the use of methylation biomarkers might significantly improve the possibility to perform early diagnosis starting from sputum.

Full table
Any biomarker to be incorporated into daily practice needs to be affordable, reproducible, and accessible to pathologists in academic and community-hospital practices and also need to be validated prospectively in the population. In this scenario, the findings of Belinsky and colleagues need further validation.
First, the correlation between increased methylation in sputum and both local and distal recurrence is quite surprising. This observation might suggest that local progression of genetic and epi-genetic alterations might reflect similar phenomena at distant sites. However, this hypothesis needs an experimental validation. Importantly, this correlation was only limited to the placebo group that included one third of the patients enrolled in the biomarker study, i.e., less than 200 subjects. Therefore, this result needs to be cautiously interpreted.
Second, the authors choose only 8 genes because, based on their previous report (7,17-19), these panel genes are cancer-specific and seems to be methylated solely in epithelial cells. A meta-analysis of thirty studies including approximately five thousand subjects screened, equally distributed between patients with lung cancer and controls and covering roughly forty different genes, has been recently published (20). This meta-analysis suggested that in addition to the genes analyzed by Belinsky and colleagues, SOX17, CDO1, ZFP42, TAC1, FAM19A4, and FHIT could be potentially considered as useful biomarkers for the screening of lung cancer. A more comprehensive approach would have probably included in the analysis a larger number of informative genes, with the hoping to improve sensitivity and specificity of the screening itself, for the detection of early stages lung cancer. Furthermore, with respect to the methodology and technology used, in the next generation sequencing (NGS) era, would be beneficial moving from a labor-intensive, gel-based technique, to a quantitative PCR technology or directly to a NGS based technology which are becoming the most widely used diagnostic tool (Table 1).
What are the implications of using these biomarkers in a screening scenario? It probably cannot be emphasized enough that the potential translation of this work would positively affect the lives of millions of people worldwide, providing to the primary care a lung cancer risk assessment test that may guide the process of decision making regarding receiving or not a CT scan in high-risk patients.
On the basis of these findings, should analysis of methylation biomarkers in liquid biopsy be incorporated into the routine molecular pathology for both the initial diagnosis and long-term surveillance in the real-world setting? Although there is still much to do with regard to the selection and standardization of methylated gene panels, we are probably moving toward to a positive conclusion.
Acknowledgements
Funding: N. Normanno is supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC) (Grant number: IG17135).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Belinsky SA, Leng S, Wu G, et al. Gene Methylation Biomarkers in Sputum and Plasma as Predictors for Lung Cancer Recurrence. Cancer Prev Res (Phila) 2017;10:635-40. [Crossref] [PubMed]
- Sugimura H, Nichols FC, Yang P, et al. Survival after recurrent nonsmall-cell lung cancer after complete pulmonary resection. Ann Thorac Surg 2007;83:409-17; discussioin 417-8.
- Koo HK, Jin SM, Lee CH, et al. Factors associated with recurrence in patients with curatively resected stage I-II lung cancer. Lung Cancer 2011;73:222-9. [Crossref] [PubMed]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484-92. [Crossref] [PubMed]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 2010;11:191-203. [Crossref] [PubMed]
- Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 2018;19:81-92. [Crossref] [PubMed]
- Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res 2006;66:3338-44. [Crossref] [PubMed]
- Herman JG, Graff JR, Myöhänen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821-6. [Crossref] [PubMed]
- Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953;6:963-8. [Crossref] [PubMed]
- Braakhuis BJ, Tabor MP, Kummer JA, et al. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res 2003;63:1727-30. [PubMed]
- Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat Rev Cancer 2018;18:19-32. [Crossref] [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Mouliere F, Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A 2015;112:3178-9. [Crossref] [PubMed]
- Yong E. Cancer biomarkers: Written in blood. Nature 2014;511:524-6. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Saccomanno G, Archer VE, Auerbach O, et al. Development of carcinoma of the lung as reflected in exfoliated cells. Cancer 1974;33:256-70. [Crossref] [PubMed]
- Leng S, Do K, Yingling CM, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res 2012;18:3387-95. [Crossref] [PubMed]
- Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 1998;95:11891-6. [Crossref] [PubMed]
- Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 2000;60:5954-8. [PubMed]
- Liu D, Peng H, Sun Q, et al. The Indirect Efficacy Comparison of DNA Methylation in Sputum for Early Screening and Auxiliary Detection of Lung Cancer: A Meta-Analysis. Int J Environ Res Public Health 2017;14:E679. [Crossref] [PubMed]


