Relationship between free and total malondialdehyde, a well-established marker of oxidative stress, in various types of human biospecimens
Introduction
Oxidative stress is a common pathophysiological pathway related to many diseases including chronic obstructive pulmonary disease (COPD), asthma, as well as the process of natural aging (1-3). It is well known that inhalation exposure to air pollution also increases levels of oxidative stress in the body (4-7). As such, there have been tremendous needs for effective biomarkers of oxidative stress in both clinical assessment of disease status and pathophysiological research. Malondialdehyde (MDA) is a product of lipid peroxidation and has been used as a biomarker of oxidative stress (8,9). It has been widely used in clinical research and environmental epidemiological studies, especially in the evaluation of oxidative stress as a response to air pollution exposure (10-13). The main endogenous production of MDA arises from the oxidation of polyunsaturated fatty acids with more than two methylene-interrupted double bonds (14,15). Besides the unconjugated form (free MDA), MDA can also exist in conjugated forms by forming covalent bonds with the -SH and -NH2 groups of proteins and nucleic acids (8,16-18). The sum of unconjugated and conjugated MDA is often termed total MDA. Previously free MDA has been more commonly used than total MDA as a biomarker of oxidative stress (10,19), perhaps due to methodological convenience as total MDA analysis requires additional hydrolysis steps. A biological specimen may contain proteins and nucleic acids that can conjugate with free MDA produced via lipid peroxidation (8,9). The measurement of total MDA, therefore, involves an acid- or base-hydrolysis at an elevated temperature to release conjugated MDA into free MDA. This hydrolysis procedure may introduce artifacts through the release of additional MDA molecules from compounds other than MDA-conjugates (8).
Because proteins and nucleic acids can scavenge free MDA, measuring free MDA may not accurately reflect the total amount of MDA generated from lipid peroxidation. On the other hand, dietary intake can contribute to conjugated MDA (8,20). Therefore, it is not clear which of the two forms (free MDA and total MDA) is a better biomarker of oxidative stress induced by lipid peroxidation. The first step towards answering this question is to understand the relationship between free and total MDA in biological specimens. To our knowledge, no studies have measured both free and total MDA within the same study to allow for a systematic evaluation.
Once produced, MDA can diffuse from its source of production into multiple biological media such as exhaled breath condensate (EBC) (12,21), nasal fluid, saliva (22,23), blood (11,24) and urine (25). EBC consists of particles or droplets aerosolized from the airway lining fluid (26) and can be collected non-invasively using a condensation device. MDA in EBC has been used as a biomarker of pulmonary oxidative stress, particularly as a response to air pollution exposure or the presence of respiratory diseases (12,21). For the evaluation of oxidative stress in the upper airway, nasal fluid can be collected using nasal lavage or an absorptive sampler (27). In saliva, the concentration of MDA has been used as an indicator of oral and systemic oxidative stress (22,23). On the other hand, MDA in blood (plasma or serum) and urine samples has been used as a biomarker of systemic oxidative stress (11,25,28). Hence, in the present study, we have examined the relationship between free MDA and total MDA in five types of commonly used human biospecimens. The results are expected to help with future researchers that plan to use MDA as a biomarker of oxidative stress.
Methods
We used banked biological samples collected in five studies (29-32). All studies had their protocols approved by an institutional review board (IRB), and all study participants provided informed consent. We received biological samples that had been de-identified and were stored in well-sealed containers at low temperature (−80 to −20 °C) before MDA analysis in our laboratory. However, the present study had no access to any individual-level subject information and detailed protocols of the original study.
Human biospecimens
In the present study, we used banked biological samples collected in five studies (Table 1, Figure S1) (29-32). All studies have had their protocol reviewed by the Ethics Committee at the institution of the study investigators. Informed consent was obtained from all study participants. Biological samples had been de-identified and were stored in well-sealed containers at low temperature (−80 to −20 °C) prior to MDA analysis in our laboratory.
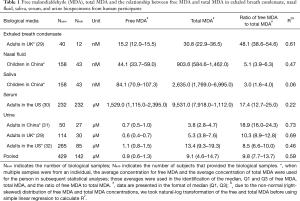
Full table
Exhaled breath condensate (EBC)
We obtained 40 EBC samples from 12 nonsmoking adults aged 45 to 88 years old living in London, UK (29). None of them were current smokers. Those study participants had either ischemic heart disease (IHD), COPD, or free from chronic diseases. The EBC samples were collected between December 2012 and June 2014 using a Jaeger EcoScreen condensing device (Jaeger, Wurzburg, Germany). After collection, the aliquots of EBC samples were stored in a −80 °C freezer and then a −20 °C freezer until analysis in September 2015.
Nasal fluid
We obtained 158 nasal fluid samples from 43 children (5–13 years old) with stable asthma. Each nasal fluid sample was collected using a mixed cellulose ester (MSE) sampling strip (HAWG047S6, Millipore Sigma, USA) in the size of 5 mm (width) × 40 mm (length). The sampling strip was inserted approximately 10 mm into the nostril. After insertion, the study participants pinched their nose with their thumb and index finger for 2 min. Both left and right nostrils were sampled simultaneously. After collection, nasal fluid strips were added with 300 µL of deionized water, vortexed for 2 min, and centrifuged for 2 min at 6,600 rpm (BSC1006-B, Benchmark Research Products, USA). The supernatant was aliquoted and stored at −20 °C, until analysis within two months after collection.
Saliva
We obtained 158 saliva samples from 43 children (5–13 years old) with stable asthma. Saliva samples were collected using the passive drooling technique. Within 30 min before saliva collection, participants were asked to refrain from eating and drinking. Immediately before saliva collection, participants were asked to rinse their mouth with distilled water. Saliva samples were stored at a −20 °C for 2 months before being transferred to a −70 °C freezer. Before sample analysis, saliva samples were thawed and centrifuged at 6,600 rpm (BSC1006-B, Benchmark Research Products, USA) for 2 min. The supernatant was used for MDA measurements, which was conducted within four months after saliva collection.
Serum
We obtained 232 serum samples from 232 Hispanic adults living in California as part of the Asthma Bio-Repository of Integrative Genetic study (30). The study participants consisted of 117 individuals with asthma and 115 individuals without asthma. Their age range was between 23 to 31 years.
Urine
We analyzed 429 banked urine samples from three studies. All study participants that provided the urine samples were not current smokers. The first study had 50 urine samples collected from 27 healthy adults (aged 22 to 27 years old) living in Beijing, China (31). The second study provided 114 urine samples collected from 30 adults (aged 45 to 88 years old) living in London, UK, who had IHD, COPD, or free from chronic diseases (29). The third study provided 265 urine samples collected from 85 adults with COPD (aged 46 to 90 years old) living in Eastern Massachusetts, USA (32).
Analysis of MDA
We used a fluorescence-generating agent, thiobarbituric acid (TBA), to derivatize MDA; and then analyzed the MDA-TBA adduct using an HPLC-fluorescence technique described previously (11). Compared to the conventional colorimetric analysis method for MDA-TBA adduct detection, the use of HPLC separation of MDA-TBA adduct in the present study minimized interference from TBA-reactive substances (TBARS) and achieves improved specificity and accuracy of MDA analysis (33).
For free MDA analysis, we started with TBA derivatization by mixing a 150 µL sample aliquot with 750 µL phosphoric acid (440 mM) and 250 µL TBA solution (42 mM). The mixture was then incubated at 80 °C for one hour to allow for the formation of MDA-TBA2. This TBA derivatization process was identical for free MDA and total MDA, while total MDA measurement used an extra step of alkaline hydrolysis before TBA derivatization. The alkaline hydrolysis procedure for the serum and urine samples involved incubating a 20 µL sample aliquot with 65 µL 1N NaOH at 60 °C for 30 min, followed by mixing it with 65 µL 1N HCl. The hydrolysis procedure for the EBC samples, however, was slightly different as follows. A 120 µL EBC aliquot was incubated with 15 µL 5N NaOH at 60 °C for 30 min, followed by adding 15 µL 5N HCl. After TBA derivatization, 20 µL of the resultant mixture was injected into the HPLC system where MDA-TBA2 adduct was isolated using a Nova-Pak C18 column (Waters, USA). The mobile phase of the HPLC system consisted of 40% (v/v) methanol and 60% (v/v) 50 mM KH2PO4 solution, its pH was adjusted to 6.8, and then the solution was filtered using a 0.22 µm nylon membrane (Catalog # 58522-N47, Microsolv Tech, USA). The flow rate of mobile phase was set at 0.8 mL/min. The MDA-TBA2 adduct was quantified using a fluorescence detector with excitation wavelength at 532 nm and emission wavelength at 553 nm. The method detection limit, extraction recovery, and analytical precision (measured as relative standard deviation from eight replicate injections) were 1.8 nM, 75.9%, and 2.2%, respectively.
Statistical analyses
When multiple samples were collected from the same study participant, the average concentrations were calculated to represent the participant. The ratio of free MDA to total MDA was calculated to assess the fraction of MDA that existed in the unconjugated form. Due to the right-skewed distribution of the free MDA concentrations, total MDA concentrations, and the ratio of free MDA to total MDA, we performed natural-log transformation before subsequent statistical analyses. Data after natural-log transformation demonstrated improved normality, indicated by histogram and the P value of Shapiro-Wilk normality test. Simple linear regression analysis was used to evaluate the relationship between free MDA and total MDA. For the comparison of MDA concentration, t-test was used to compare means between the two groups. One-way analysis of variance (ANOVA) was used to compare the natural log of the ratio of free MDA to total MDA across various types of biospecimens in combination with Tukey’s test for post-hoc pairwise comparisons. All statistical analyses were conducted using the R (Version 3.4.0) (34).
Results
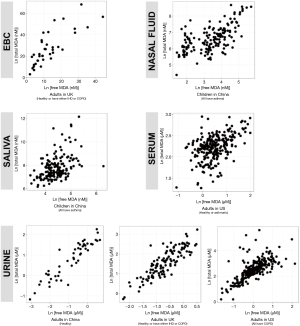
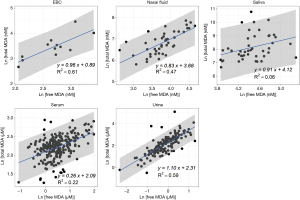
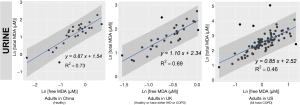
We reported summary statistics of free MDA, total MDA, and the relationship between free MDA and total MDA in various types of biospecimens in Table 1. The median of the ratio of free MDA to total MDA was 48.1% in EBC, 17.4% in serum, 9.8% in urine samples, 5.1% in nasal fluid, and 3.0% in saliva. According to one-way ANOVA and post-hoc pairwise comparison by Tukey’s test, the ratio of free MDA to total MDA in EBC > serum > urine > nasal fluid > saliva (P<0.001). The coefficient of determination (R2) of linear regression between the natural log of free MDA and the natural log of total MDA was 0.61 in EBC, 0.47 in nasal fluid, 0.06 in saliva, 0.22 in serum, and 0.59 in urine samples (Table 1, Figure 1). The R2 values were similar for the urines samples collected by two studies (R2=0.73 and 0.69, respectively), but was lower in the third study (R2=0.46) (Figure 2).
Discussion
Both free MDA and total MDA have been used as a biomarker of oxidative stress (11,21,25,35,36). In this study, we used hydrolysis to release conjugated MDA to measure total MDA in addition to the assessment of free MDA in the biological specimens collected. All sample analysis was performed following the same protocol to facilitate comparability. Although the conventional spectrophotometric assay was a commonly used method of MDA quantification, we did not use it in our study. The conventional spectrophotometric assay evaluates the absorbance or fluoresces of the MDA-TBA adduct directly without any specific separation steps. Thus it cannot discern the MDA-TBA adduct from TBA adducts with other TBA-reactive substances that exhibit absorbance or fluoresces at the same wavelengths as MDA-TBA. This can introduce additional “false positive” reading for MDA quantification (8,37). In contrast to this method, we used HPLC before fluorescence quantification to separate MDA-TBA from TBA adducts with other TBA-reactive compounds, to achieve a higher specificity for MDA assessment.
EBC is highly dilute and has a low-protein aqueous matrix. The protein and protein adducts in airway epithelial lining fluid have relatively large molecular weight. Thus it is relatively difficult for them to be collected as a constituent of EBC (26,38). In the present study, we observed a high fraction of unconjugated MDA in total MDA (i.e., free MDA to total MDA ratio) in EBC (median =48.1%), which was the highest among all biological samples examined in this study. The concentrations of free MDA and total MDA in EBC were significantly correlated with each other, and the relationship between ln(free MDA) and ln(total MDA) had an R2 =0.61. These findings are consistent with existing studies, which reported a low level of MDA-reactive substances (such as protein) in EBC (8,9,16,26). This lack of MDA-reactive substances limited the formation of MDA adducts and thus may contribute to the high percentage of free MDA in total MDA, as well as the linear relationship between free MDA and total MDA in EBC samples examined in this study. Overall, our findings suggest that using either free MDA or total MDA in EBC may not make a difference in reflecting oxidative stress levels, given the high correlations between the two forms of MDA.
Previous studies have collected nasal secretions to gain insight into nasal pathophysiology, mainly the evaluation of mucosal inflammation by the measurement of cytokines and inflammatory mediators (27,39). To our knowledge, there have been no published studies that investigated the level of free MDA or total MDA in nasal fluids. In our study, we observed that the percentage of total MDA that existed as free MDA in nasal fluid samples was relatively low (median =5.1%). The R2 of the linear regression between ln(free MDA) and ln(total MDA) was 0.47, indicating they were not linearly correlated. Previous studies have detected a considerable variety of proteins in nasal fluid, including mucus glycoproteins, interleukin (IL)-1β, IL-8, tumor necrosis factor-α, eosinophil cationic protein, and tryptase (27,39,40). As MDA can form adducts with proteins (8), it is understandable that most of the MDA in nasal fluid samples examined in this study existed in a conjugated form. The varying level of nasal mucosal inflammation caused by stimulation from bacteria, virus, allergen and other irritants in inhaled air may result in varying concentrations of proteins in nasal fluid samples (27,39), thus contributing to the observed poor correlation between free MDA and total MDA in nasal fluid. Due to the low correlations between free and total MDA in nasal fluid, whether free MDA or total MDA better reflects oxidative stress levels needs to be evaluated in future studies.
Despite the traditional perception of saliva as a digestive fluid where its endogenous enzymes help break down lipids and starch, studies in recent years have found that biomarkers in saliva can reflect oral and systemic health conditions (41,42). Previous studies have measured MDA in saliva as indicators of oxidative stress in individuals with oral diseases (such as chronic periodontitis) and metabolic diseases (such as diabetes) (22,23). In our study, the saliva samples demonstrated the lowest ratio of free MDA to total MDA in all five types of biospecimens examined. The R2 between ln(free MDA) and ln(total MDA) was very low (0.06). This is consistent with the complex composition of saliva. Previous studies reported that saliva contains glycoproteins that protect oral health, including the salivary mucins and proline-rich glycoproteins that facilitate remineralization and resist demineralization (43). Inflammatory cytokines have also been detected in saliva, including IL-1β, IL-6, IL-8 and tumor necrosis factor (42). The presence of those proteins facilitates the formation of MDA adducts, thus resulting in the high proportion of conjugated form of MDA in salivary MDA. Variation in the concentration of those proteins may have contributed to low correlations between free MDA and total MDA in saliva. In addition, although the study participants rinsed their mouth with deionized water before saliva collection, it is possible that there might be some food residues in saliva. Most MDA in food is in the conjugated form (8), this might be another factor that contributes to the high proportion of conjugated form of MDA in saliva as observed in our study. As free and total MDA exhibited low correlations in saliva, which one better reflects the levels of oxidative stress remains to be evaluated in future studies.
In serum samples examined in this study, the percentage of total MDA that existed as free MDA was relatively low (17.4%, see Table 1 and Figure 3). This is consistent with the observation by other researchers that free MDA can react extensively with serum albumin in cell culture medium (44). Previous studies in healthy individuals also reported that the binding of MDA with -NH2 group of proteins accounted for 83.2% of total MDA in serum, and 83.5% to 92% of total MDA in plasma, respectively (45,46). A study by De Vecchi et al. [2009] compared end-stage renal disease patients to healthy individuals and found that plasma level of total MDA and bound MDA can be affected by both renal clearance and albumin concentrations (47). In this study, the relatively low R2 value (R2=0.22) between serum free MDA and total MDA may be the result of inter-person differences in serum albumin concentration and renal clearance of MDA (47). As the correlations between free and total MDA were low in the serum samples examined in this study, whether free or total MDA in serum better reflects systemic oxidative stress levels needs to be evaluated in future studies.
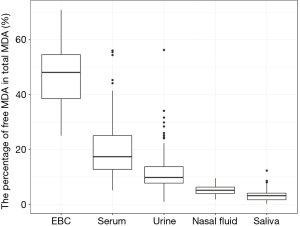
The sources of urinary MDA included dietary consumption of food that contained MDA adducts with proteins and endogenous production of MDA from lipid peroxidation and prostaglandin metabolism (14,48,49). In the present study, the percentage of total MDA that existed as free MDA was the lowest in urine compared to EBC and serum (P<0.001). This was consistent with the finding of previous studies where free MDA typically constituted only a small proportion of urinary MDA. The majority of urinary MDA exists in the form of adducts with lysine, N-acetylated derivative of lysine, phospholipid bases (serine and ethanolamine) and the nucleic acid bases (guanine and deoxyguanosine) (8,20,50). Though dietary intake contributes to a significant portion of urinary MDA, increases in total MDA and free MDA in urine have been associated with an increase in oxidative stress in vivo (50).
Despite the relatively low proportion of free MDA in urinary MDA, the concentration of urinary free MDA was significantly correlated with urinary total MDA in all three studies examined. In most urine samples examined in this study, the relationship between the natural-log of free MDA and the natural log of total MDA was moderately linear. This might be due to the relatively consistent concentration of amino acids and nucleic acids that are typically present in urine samples. Among individuals with uncompromised glomerular filtration and renal tubule function, more than 99% of amino acid and nucleic acids are reabsorbed in the proximal tubule, leaving only ~1% in the final urine (51). This consistent amount of amino acid and nucleic acids excreted into the final urine might contribute to the relatively consistent percentage of bound MDA in total MDA, thereby resulting in the moderately linear relationship between the natural log of free MDA and the natural log of total MDA. The correlation between free and total MDA was observed to be high (R2=0.73) in urine samples from healthy study participants in China but much lower among the COPD patients in the US (R2=0.46). It was beyond the scope of the current study to disentangle the effect of locality and disease status on the correlation between free and total MDA in urine samples. Further studies would be required to address whether the correlation between free and total MDA in urine varies by disease status.
Our study had certain limitations. First, the present study is descriptive in nature. We utilized existing banked biospecimens originally collected in several studies with distinct aims. This limits our ability to examine the relationships between free and total MDA in a more mechanistic fashion. Second, these “convenient” samples used in the present study do not necessarily represent the general population where the study subjects came from. Third, we did not compare the performance of free and total MDA with a “gold standard” biomarker for oxidative stress, such as the production rate of reactive oxygen species (10). However, the knowledge of the relationship between free and total MDA in different biological media can serve as the first step towards a better understanding of selecting free MDA or total MDA as an indicator of oxidative stress. For future studies that investigate free MDA and total MDA as biomarkers of lipid peroxidation, it is also important to keep in mind that unbound MDA is a toxic molecule that possesses mutagenic and atherogenic potential due to its ability to form adducts with nucleic acids and proteins (14,15,20). Hence, the levels of free MDA and total MDA in a biological specimen and its role in downstream toxicological pathways should be taken into consideration.
Conclusions
For exhaled breath condensate and urine samples, using either free MDA or total MDA as a biomarker for oxidative stress may not make a difference given high correlations between the two forms of MDA. Given the observed low correlation between free and total MDA in the nasal fluid, saliva, and serum, further studies are required to elucidate whether the free or total MDA in those specimens best represent oxidative stress. In addition, future research is needed to understand the effect of disease status on the correlation between free and total MDA.
Acknowledgements
We would want to acknowledge Dr. Yan Chang (formerly at University of Southern California) for her assistance with total MDA method development.
Funding: This study was supported by a graduate fellowship to Cui from Duke University Integrated Toxicology and Environmental Health Program and Duke University Graduate School. The funding support for this study also include a grant from British Heart Foundation (PGF/10/82/28608), a grant from the Victor Phillip Dahdaleh Charitable Foundation, a grant from the Hastings Foundation, a research grant from Underwriters Laboratories, Inc., a grant from the National Heart, Lung and Blood Institute (R01HL061768), and grants from National Institute of Environmental Health Sciences (R01ES019853, R01ES021801, R01ES023262, R01ES025786, P30ES007048). In addition, the research work of this study is supported by the Imperial College/Kings College MRC-PHE for Environment and Health and the National Institute for Health Research (NIHR), Respiratory Disease Biomedical Research Unit at the Royal Brompton NHS Foundation Trust and Imperial College London, and by resources and the use of facilities at the VA Boston Healthcare System.
Footnote
Conflicts of Interest: Zhang is a co-investigator of another study funded by the Underwriters Laboratories, Inc on the health impact of indoor air filtration in asthmatic children. He also received an honorarium for attending a global advisory board meeting on air pollution from the RB Company in London, UK. Chung has received honoraria for participating in Advisory Board meetings regarding treatments for asthma and COPD for GlaxoSmithKline, Boehringer Ingelheim, AstraZeneca, Novartis, and Johnson & Johnson, and has received grant funding through his institution from Pfizer, GlaxoSmithKline, and Merck. He has also been remunerated for speaking engagements by AstraZeneca, Merck, and Novartis. The other authors have no conflicts of interest to declare.
Ethical Statement: All studies had their protocols approved by an institutional review board (IRB), and all study participants provided informed consent.
References
- Kirkham PA, Barnes PJ. Oxidative stress in COPD. Chest 2013;144:266-73. [Crossref] [PubMed]
- Kirkham P, Rahman I. Oxidative stress in asthma and COPD: Antioxidants as a therapeutic strategy. Pharmacol Ther 2006;111:476-94. [Crossref] [PubMed]
- Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol 2007;292:R18-36. [Crossref] [PubMed]
- Kelly FJ. Oxidative stress: its role in air pollution and adverse health effects. Occup Environ Med 2003;60:612-6. [Crossref] [PubMed]
- Mudway IS, Kelly FJ. Ozone and the lung: a sensitive issue. Mol Aspects Med 2000;21:1-48. [Crossref] [PubMed]
- Schlesinger RB, Kunzli N, Hidy GM, et al. The health relevance of ambient particulate matter characteristics: coherence of toxicological and epidemiological inferences. Inhal Toxicol 2006;18:95-125. [Crossref] [PubMed]
- Romieu I, Castro-Giner F, Kunzli N, et al. Air pollution, oxidative stress and dietary supplementation: a review. Eur Respir J 2008;31:179-97. [Crossref] [PubMed]
- Draper HH, Hadley M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol 1990;186:421-31. [Crossref] [PubMed]
- Hageman JJ, Bast A, Vermeulen NP. Monitoring of oxidative free radical damage in vivo: analytical aspects. Chem Biol Interact 1992;82:243-93. [Crossref] [PubMed]
- Ho E, Karimi Galougahi K, Liu CC, et al. Biological markers of oxidative stress: Applications to cardiovascular research and practice. Redox Biology 2013;1:483-91. [Crossref] [PubMed]
- Nielsen F, Mikkelsen BB, Nielsen JB, et al. Plasma malondialdehyde as biomarker for oxidative stress: reference interval and effects of life-style factors. Clin Chem 1997;43:1209-14. [PubMed]
- Gong J, Zhu T, Kipen H, et al. Malondialdehyde in exhaled breath condensate and urine as a biomarker of air pollution induced oxidative stress. J Expo Sci Environ Epidemiol 2013;23:322-7. [Crossref] [PubMed]
- Romieu I, Barraza-Villarreal A, Escamilla-Nunez C, et al. Exhaled breath malondialdehyde as a marker of effect of exposure to air pollution in children with asthma. J Allergy Clin Immunol 2008;121:903-9.e6. [Crossref] [PubMed]
- Draper HH, McGirr LG, Hadley M. The metabolism of malondialdehyde. Lipids 1986;21:305-7. [Crossref] [PubMed]
- Del Rio D, Stewart AJ, Pellegrini N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr Metab Cardiovasc Dis 2005;15:316-28. [Crossref] [PubMed]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 1991;11:81-128. [Crossref] [PubMed]
- Mooradian AD, Lung CC, Pinnas JL. Glycosylation enhances malondialdehyde binding to proteins. Free Radic Biol Med 1996;21:699-701. [Crossref] [PubMed]
- Agarwal S, Wee JJ, Hadley M, et al. Identification of a deoxyguanosine-malondialdehyde adduct in rat and human urine. Lipids 1994;29:429-32. [Crossref] [PubMed]
- Ayala A, Munoz MF, Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev 2014;2014:360438. [Crossref] [PubMed]
- Draper HH, Hadley M. A review of recent studies on the metabolism of exogenous and endogenous malondialdehyde. Xenobiotica 1990;20:901-7. [Crossref] [PubMed]
- Bartoli ML, Novelli F, Costa F, et al. Malondialdehyde in Exhaled Breath Condensate as a Marker of Oxidative Stress in Different Pulmonary Diseases. Mediators Inflamm 2011;2011:891752. [Crossref] [PubMed]
- Smriti K, Pai KM, Ravindranath V, et al. Role of salivary malondialdehyde in assessment of oxidative stress among diabetics. J Oral Biol Craniofac Res 2016;6:41-4. [Crossref] [PubMed]
- Khalili J, Biloklytska HF. Salivary malondialdehyde levels in clinically healthy and periodontal diseased individuals. Oral Dis 2008;14:754-60. [Crossref] [PubMed]
- Tucker PS, Dalbo VJ, Han T, et al. Clinical and research markers of oxidative stress in chronic kidney disease. Biomarkers 2013;18:103-15. [Crossref] [PubMed]
- Guichardant M, Valette-Talbi L, Cavadini C, et al. Malondialdehyde measurement in urine. J Chromatogr B Biomed Appl 1994;655:112-6. [Crossref] [PubMed]
- Hunt J. Exhaled Breath Condensate—an overview. Immunol Allergy Clin North Am 2007;27:587-96. v. [Crossref] [PubMed]
- Riechelmann H, Deutschle T, Friemel E, et al. Biological markers in nasal secretions. Eur Respir J 2003;21:600-5. [Crossref] [PubMed]
- Farouk A, Hassan MH, Nady MA, et al. Role of oxidative stress and outcome of various surgical approaches among patients with bullous lung disease candidate for surgical interference. J Thorac Dis 2016;8:2936-41. [Crossref] [PubMed]
- Sinharay R, Gong J, Barratt B, et al. Respiratory and cardiovascular responses to walking down a traffic-polluted road compared with walking in a traffic-free area in participants aged 60 years and older with chronic lung or heart disease and age-matched healthy controls: a randomised, crossover study. Lancet 2018;391:339-49. [Crossref] [PubMed]
- Breton CV, Siegmund KD, Joubert BR, et al. Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS One 2014;9:e99716. [Crossref] [PubMed]
- Rich DQ, Kipen HM, Huang W, et al. Association between changes in air pollution levels during the beijing olympics and biomarkers of inflammation and thrombosis in healthy young adults. JAMA 2012;307:2068-78. [Crossref] [PubMed]
- Grady ST, Koutrakis P, Hart JE, et al. Indoor black carbon of outdoor origin and oxidative stress biomarkers in patients with chronic obstructive pulmonary disease. Environ Int 2018;115:188-95. [Crossref] [PubMed]
- Moselhy HF, Reid RG, Yousef S, et al. A specific, accurate, and sensitive measure of total plasma malondialdehyde by HPLC. J Lipid Res 2013;54:852-8. [Crossref] [PubMed]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017.
- Schock BC, Young IS, Brown V, et al. Antioxidants and oxidative stress in BAL fluid of atopic asthmatic children. Pediatr Res 2003;53:375-81. [Crossref] [PubMed]
- Goncalves MS, Fabris BA, Brinholi FF, et al. Increased oxidative stress in foam cells obtained from hemodialysis patients. Hemodial Int 2013;17:266-74. [Crossref] [PubMed]
- Knight JA, Pieper RK, McClellan L. Specificity of the thiobarbituric acid reaction: its use in studies of lipid peroxidation. Clin Chem 1988;34:2433-8. [PubMed]
- Griese M, Noss J, von Bredow C. Protein pattern of exhaled breath condensate and saliva. Proteomics 2002;2:690-6. [Crossref] [PubMed]
- Watelet JB, Gevaert P, Holtappels G, et al. Collection of nasal secretions for immunological analysis. Eur Arch Otorhinolaryngol 2004;261:242-6. [Crossref] [PubMed]
- Patow CA, Shelhamer J, Marom Z, et al. Analysis of human nasal mucous glycoproteins. Am J Otolaryngol 1984;5:334-43. [Crossref] [PubMed]
- Yoshizawa JM, Schafer CA, Schafer JJ, et al. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev 2013;26:781-91. [Crossref] [PubMed]
- Rathnayake N, Åkerman S, Klinge B, et al. Salivary biomarkers for detection of systemic diseases. PLoS One 2013;8:e61356. [Crossref] [PubMed]
- Van Nieuw Amerongen A, Bolscher JG, Veerman EC. Salivary proteins: protective and diagnostic value in cariology? Caries Res 2004;38:247-53. [Crossref] [PubMed]
- Bird RP, Draper HH. Uptake and oxidation of malonaldehyde by cultured mammalian cells. Lipids 1982;17:519-23. [Crossref] [PubMed]
- Carbonneau MA, Peuchant E, Sess D, et al. Free and bound malondialdehyde measured as thiobarbituric acid adduct by HPLC in serum and plasma. Clin Chem 1991;37:1423-9. [PubMed]
- Hong YL, Yeh SL, Chang CY, et al. Total plasma malondialdehyde levels in 16 Taiwanese college students determined by various thiobarbituric acid tests and an improved high-performance liquid chromatography-based method. Clin Biochem 2000;33:619-25. [Crossref] [PubMed]
- De Vecchi AF, Bamonti F, Novembrino C, et al. Free and total plasma malondialdehyde in chronic renal insufficiency and in dialysis patients. Nephrol Dial Transplant 2009;24:2524-9. [Crossref] [PubMed]
- Draper HH, Hadley M, Lissemore L, et al. Identification of N-epsilon-(2-propenal)lysine as a major urinary metabolite of malondialdehyde. Lipids 1988;23:626-8. [Crossref] [PubMed]
- Pryor WA, Stanley JP. Letter: A suggested mechanism for the production of malonaldehyde during the autoxidation of polyunsaturated fatty acids. Nonenzymatic production of prostaglandin endoperoxides during autoxidation. J Org Chem 1975;40:3615-7. [Crossref] [PubMed]
- Draper HH, Csallany AS, Hadley M. Urinary aldehydes as indicators of lipid peroxidation in vivo. Free Radic Biol Med 2000;29:1071-7. [Crossref] [PubMed]
- Boron WF, Boulpaep EL. Medical physiology: a cellular and molecular approach. Philadelphia, PA: Saunders/Elsevier, 2009.


