Circulating tumor cells (CTCs) for the noninvasive monitoring and personalization of non-small cell lung cancer (NSCLC) therapies
Introduction
The development of precision medicine for cancer patients is highly dependent on the identification of the molecular drivers of their disease. Today still therapy decisions are mainly performed on the basis of tumor biopsies. Because of the widespread intratumor heterogeneity, a single biopsy is rarely able to capture the full spectrum of molecular alterations present within a tumor. To identify resistance mutations at disease progression and re-evaluate the changing gene profile of tumors the serial biopsies are required. Subjecting patients to serial biopsies however are complicated, invasive and in most cases not performed. Liquid biopsy offers a non-invasive complement to tumor biopsy for the assessment of the tumor mutational status and monitoring genetic changes in real-time. Nowadays the analysis of circulating tumor cells (CTCs) and/or circulating tumor DNA (ctDNA) opens new possibilities for cancer patient management. CTCs have the potential to provide complementary information to that delivered by even multiple tumor biopsies. Because they are shed from primary tumor and/or spatially distinct metastatic regions, CTCs provide a global representation of tumor genetic diversity. In contrast to ctDNA, CTCs might be analyzed at single cell level, thus providing a matchless representation of tumor heterogeneity and clonal dominance in a real-time. Moreover, CTCs can be engrafted into mice, in order to obtain preclinical models. CTCs are the main route of metastatic dissemination and contain subpopulations with tumor-initiating potential. Thus, advances in CTC molecular and functional characterization increase our knowledge about the metastasis formation mechanisms, and the combination of CTC and ctDNA assays through routine blood test would provide biomarkers for the real-time patient monitoring and therapy stratification, in different tumor types including non-small cell lung cancer (NSCLC).
Molecular profiles of patients
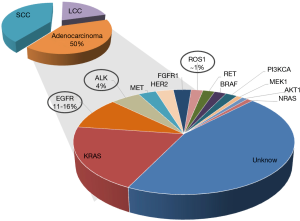
At initial diagnosis a majority of NSCLC patients present advanced disease for which no curative therapy exists. NSCLC and in particular adenocarcinoma have been classified into clinically relevant molecular subsets according to multiple oncogenic driver alterations (Figure 1) (1). These alterations activate prosurvival oncogenic signaling pathways promoting the tumor growth.
Epidermal growth factor receptor (EGFR) mutations
Common EGFR alterations (exon 19 deletions and the L858R point mutation) are identified in 11 to 16% of patients with NSCLC in Western countries and in about 50% of Asian patients and confer sensitivity to EGFR tyrosine kinase inhibitor (TKI) therapy such as erlotinib, gefitinib, or afatinib. The most frequent EGFR mutations affect the EGFR kinase catalytic domain and TKIs are standard first-line therapy that is associated with superior radiographic response and prolonged progression free survival (PFS) compared to standard cytotoxic chemotherapy (2,3). The most frequent EGFR alterations, namely deletions within exon 19 and L858R point mutation account for more than 90% of EGFR mutated patients. The response to EGFR TKI is positively related to constitutional activation of EGFR signaling, which has been shown to be particularly high in non-smoker patients with lung adenocarcinoma. Despite the impressive efficacy of the TKI therapy, most patients will develop clinical evidence of acquired resistance to treatment after a median of 12 months (2,3). Resistance to TKIs is generally classified as either primary (i.e., intrinsic) or secondary (i.e., acquired). Less is known about the basis of intrinsic resistance, which is defined as a de novo lack of treatment response (4). As commonly observed in oncogene-driven malignancies, acquired resistance to EGFR inhibitors is divided into two main groups: on-target mechanisms such as the acquisition of additional genetic alteration in the drug target itself through secondary mutations or gene amplification, changes in tumor histology or alterations in drug metabolism (4). The principal T790M resistance mutation is detectable in approximately 50% of TKI-resistant patients. In rare cases it has been also identified in tumors before treatment with EGFR TKI. Three additional secondary mutations have been associated with TKI resistance (D761Y, T854A, L747S) and C797S was described as mutation, responsible for acquired resistance to osimertinib (third generation EGFR TKI) (5). Several bypass pathways implicated in EGFR TKI resistance include MET, HER2, FGFR and AXL amplification, and acquired mutations in other oncogenes such as PIK3CA and BRAF. Third generation EGFR TKI—osimertinib, which targets both, activating EGFR gene alterations and the T790M mutation is now the standard front-line therapy for EGFR-mutant NSCLC. Up to now the identification of EGFR mutations is mainly performed on tumor biopsies, which carry risks, linked to the chirurgical intervention itself and could be not feasible, in particular at disease progression. Liquid biopsies comprising CTCs and circulating tumor DNA (ctDNA) enable such mutation analysis for diagnosis and monitoring over the course of the therapy.
Anaplastic lymphoma kinase (ALK)- and c-ros oncogene 1 (ROS1)-rearrangements
A unique molecular subgroup of 4% of NSCLC patients presents the ALK gene rearrangements, which involve the ALK gene and, most often, the echinoderm microtubule-associated protein-like 4 (EML4) locus in opposite directions (6). Several different in-frame fusion variants of EML4-ALK have been described with different EML4 breakpoints. However, all fusion variants harbor identical C-terminal domain conferring a gain of function leading to constitutively active fusion protein with robust transforming activity. In 2012, on the basis of the response rates demonstrated in phase I and phase II studies, crizotinib obtained FDA approval to be used as a second line therapy for advanced ALK-rearranged NSCLC (7,8) patients. It is now the standard therapy in advanced, already treated ALK-rearranged patients, as well in the first line setting (9,10). Another molecular driver event also sensitive to TKI is ROS1 fusion gene present in about 1% of NSCLC patients that could benefit from crizotinib treatment. Although the TKI show clinical efficacy with significantly prolonged progression-free survival, most of the patients that benefit from TKI treatment, similarly to EGFR mutated patients, will inevitably develop acquired resistance.
ALK resistance mechanisms have been identified in ~30–40% of crizotinib-resistant patients (11). The “gatekeeper” L1196M and G1296A (12) are the most often identified secondary ALK resistance mutations. Additional secondary mutations detected in crizotinib-resistant ALK-positive tumors are localized throughout the ALK kinase domain, they include: 1151Tins, F1174C/L/V, L1152R, E1210K, C1156Y, I1171T, S1206Y, V1180L (13,14). The study reported that cell lines established from biopsies of patients with crizotinib-resistant NSCLC harboring some of these mutations including the most frequent L1196M and G1296A were sensitive to ceritinib (15). For patients relapsing on crizotinib, more potent second-generation ALK inhibitors such as ceritinib, alectinib and brigatinib, have become standard treatments, re- inducing responses to treatment in the majority of crizotinib-resistant patients. The F1174C/L/V, 1151Tins, L1152P, C1156Y and G1202R mutations have been associated with resistance to ceritinib. Resistance to alectinib was found after acquiring V1180L and I1171T/N/S mutations (16) and double mutations in E1210K and S1206C or D1203N confer resistance to brigatinib (17). Mutations in ALK kinase domain emerging on treatment with second-generation ALK inhibitors are only targetable by lorlatinib (third generation TKI). However recently has been described the re-sensitization to crizotinib in the tumor that acquired both the C1156Y and L1198F mutations during the lorlatinib treatment (18). In approximately one-third of crizotinib-resistant tumors, the activation of by-pass signaling pathways, such as the development of EGFR mutations or activation of wild-type EGFR, KIT or HER2, has been identified as a cause of acquired resistance [4]. In about 30% of patients the resistance mechanism is still unidentified (19).
Targeting
Recent clinical studies are giving growing arguments for the using next-generation TKI as upfront therapeutic option in ALK and EGFR NSCLC. Indeed nowadays according to encouraging results from the clinical studies upfront alectinib (ALK patients) or osimertinib (in EGFR cases) show greater efficacy in the treatment of brain metastases, less toxicity and prolonged PFS than the first generation TKI (20,21). Ongoing are clinical trials with the frontline lorlatinib treatment for ALK rearranged tumors (22,23).
Importance of non-invasive routine monitoring
Selective TKIs have dramatically transformed the therapeutic landscape for NSCLC. However, acquired resistance to targeted therapies remains a persistent issue, and the development of resistant cell subpopulations within highly heterogeneous cancers still displays an important obstacle to the effective cancer treatment.
Novel biomarkers are, therefore, needed to aid in clinical decision making and ultimately improve patient’s outcomes. The prognostic biomarkers are able to state and inform about the cancer features and evolution. Another group of biomarkers has the potential to monitor therapeutic responses and predict recurrences through serial sampling. The CTCs enumeration represents a well-established prognostic value for several tumor types. Recently CTCs are being investigated in parallel to ctDNA and microRNA as a candidate predictive biomarker in different tumors, between them also for NSCLC monitoring.
Tumor biopsies may not represent the wide spectrum of genetic aberrations present within a tumor. Because CTCs may be shed from distinct metastatic sites, they are likely more representative of the heterogeneous nature of tumors. The tremendous spatial and temporal heterogeneity in primary and metastatic tumors has been contemporized recently owing to advances in next generation sequencing technologies. However application of such analyses to routine clinical applications is burdened with high technical and medical risks, particularly for cancers like NSCLC that frequently metastasize to the brain, lungs or bones. Finally repeated biopsies from progressing tumors are risky from the medical point of view and the routine sampling of tumors at several time points of treatment remains not only a technical challenge but also a controversial issue. Thus the population of CTCs isolated from the patients’ blood provides a much more easily accessible source of tumor material which in addition may be representative of disseminated cancer cells derived from various metastatic sites within individual patient.
CTC as unique source of information
Noninvasive peripheral blood sampling offers the possibility to obtain a subpopulation of disseminated tumor cells that are the main route of metastatic progression and may contain tumorigenic clones (24). Although the existence of CTCs in blood was first reported in 1869 (25), the CTCs’ isolation is still challenging because of their rarity, fragility and heterogeneity. Only recent advances in technologies, although consuming and technologically difficult gave the possibility to efficiently isolate rare CTCs, potentially enabling the study of tumor heterogeneity and treatment resistance mechanisms at the single CTC level. The enumeration of CTCs is nowadays of strong prognostic value in several cancer types and can be applied clinically as a pharmacodynamic biomarker. Indeed multifaceted CTC-based technologies are expanding into a wide spectrum of tools with a matchless interest in precision oncology, they include: role of predictive biomarkers, understanding the mechanisms and sources of tumor heterogeneity and metastasis but also personalized testing of drug efficacy.
Besides CTCs, the analysis of ctDNA is increasingly used to overcome tumor biopsy limitations. Moreover, circulating microRNAs, extracellular vesicles and tumor-educated platelets are starting to complement the liquid biopsy with potential forthcoming clinical applicability (26,27). Using the ctDNA to monitor the emergence of mutations during cancer progression possesses certain restrictions since the tumor evolution arises not only through genetic changes, but also from phenotypic alterations through epigenetic reprograming. Such functional modifications require the analysis of living cells. In addition CTCs analysis may constitute a unique approach to properly analyze tumor heterogeneity at the single cell level. Few recent works nicely show the parallel analysis of CTCs and ctDNA as a complement characteristic of tumor evolution and possibility to guide future management of the patients. Although radiographically directed needle biopsies of recurrent or metastatic tumors are still current, clinical standard for EGFR genotyping in the setting of acquired resistance, some laboratories started to use the supplementary analysis of simultaneously collected CTC and ctDNA. Indeed Punnoose et al. (28) detected EGFR mutations in CTC and ctDNA, which were strongly concordant with mutation status in matched tumor. Maheswaran et al. (29) described the detection of expected EGFR-activating mutations in CTCs from 11 of 12 tested NSCLC patients. Additionally, the T790M resistance mutation has been found in the CTCs of patients who had already received TKI. The CTCs have been isolated for this study using CTC-chip, a microfluidic equipment comprising micoparticules coated with anti-EpCAM antibodies. Sundaresan et al. (30) show also a successful T790M genotyping in CTCs and ctDNA of NSCLC patients. Discrepancies between a single tumor biopsy and blood-based sampling may result at least in part from the fact that the latter likely includes material from multiple disease sites. In the light of those promising reports the molecular analysis of CTCs supplemented with ctDNA will certainly contributes to reveal the characteristic of cancer cell dissemination and identify signaling pathways and potentially targetable genes for therapeutic interventions. CTCs offer greater possibilities of analysis (Figure 2), behind DNA evaluation, like RNA and proteomic analysis, epigenetic profiling, functional studies and development of CTC derived xenograft (CDX) models, all of those requires integrated and alive cell.
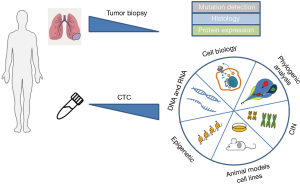
CTC ongoing and future advantages
No universal technology for CTC detection and evaluation
CTCs are rare, occurring at a rate of one cell per 106 or 107 leucocytes in the human blood. The combination of two successive steps: (I) an initial enrichment and (II) CTC detection by different phenotypical or genetic techniques, are required for reliable CTCs identification. Various technical efforts have been undertaken to properly detect and quantify CTCs, although the elaboration of a universal assay is still pretty debatable. The rarity and molecular and phenotypical heterogeneity of CTCs make their detection highly difficult and although multiple technologies have been developed, there is no consensus on their choice. The method of CTC analysis should rather be chosen according to the question asked (for example enumeration in a clinical setting or identification of predictive biomarkers or single cell molecular analysis). CellSearch platform uses the cell surface epithelial cell adhesion molecule (EpCAM) to enrich epithelial CTCs. In this platform, enriched cells are identified as CTCs by the costaining with anti-cytokeratin (CK) antibodies, and potentially contaminating PBMC are detected by CD45 counterstain. This system represents the current gold standard for CTC enumeration and is approved by the US Food and Drug Administration (FDA) as an aid to prognosis for patients with metastatic breast, colorectal, and prostate cancer. EpCAM dependence in this method may occur disadvantageous for the CTCs, which have already lost their epithelial characteristic and express epithelial-mesenchymal transition (EMT) markers. Those CTCs undergoing EMT may be missed by the CellSearch (31). The initial report about the feasibility of the enumeration of CTCs in the blood of NSCLC patients using the CellSearch (32) has been followed by the report of Krebs et al. (33) that for the first time demonstrated the prevalence and prognostic significance of CTCs in patients with NSCLC. A cutoff value of >5 CTCs in 7.5 mL of blood was highly significant in predicting worse prognosis comparing to patients having fewer than 5 CTCs (33). Few others clinical reports including our own confirmed the prognostic value of CTCs in NSCLC patients at the baseline (28,34-36).
Using an enrichment method based on blood filtration (ISET, Isolation by Size Epithelial Tumor cells), the prognostic significance of CTCs in NSCLC patients was also stated by Hofman et al. (37). We and others demonstrated that the ISET approach identifies a higher proportion of CTC-positive NSCLC patients than CellSearch equally stating high sensitivity for CTC enrichment from blood samples using the ISET technique in the case of NSCLC patients (33,38,39). NSCLC CTCs were identified in higher numbers by ISET compared to the CellSearch platform most likely because CTCs that lost epithelial features are missed by CellSearch (31,40). ISET technique allows conserving and analyzing CTCs posteriori that is particularly useful for longitudinal research. In spite of technical difficulties, the usability of CTC assays as predictive biomarkers has been demonstrated in a few reports. Indeed the differences in the expression of EMT markers may depend on the driver gene/mutation in NSCLCs and potentially confer the predictive factor (41). Two groups including our own have initially reported the feasibility of ALK-rearrangement detection in CTCs enriched by ISET in ALK positive NSCLC patients (42,43). Our group developed a FISH (Fluorescent In Situ Hybridization) technique for CTCs enriched on filters [filter-adapted FISH (FA-FISH)]. Using a FA-FISH, we showed that ALK-rearranged CTCs might be identified in a cohort of 18 ALK- positive NSCLC patients. 100% of tested ALK-positive patients had four or more ALK-rearranged CTCs per 1 mL of blood, while all of ALK-negative patients presented none or one CTC with ALK-rearrangement signal. Interstingly ALK-rearranged CTCs presented a unique ALK rearrangement pattern. Additionally, our group tested markers of EMT in ALK-rearranged CTCs. We establish that CKs and E-cadherin were not detected in ALK-rearranged CTCs, contrary to mesenchymal markers (vimentin, N-cadherin), that were exclusively expressed. Indeed, ALK-rearranged CTCs displayed a mesenchymal phenotype while epithelial and mesenchymal markers are detected in heterogeneous tumors from corresponding patient (41). These data suggest that CTCs may harbor migratory and invasive properties in these patients. Ilie et al. showed the detection of ALK rearrangement accompanied with robust ALK protein expression by immunohistochemistry in CTCs from five ALK-rearranged NSCLCs. The ALK-rearrangement status in CTCs at the baseline and after few systemic treatments has been reported also by Provencio et al. (43) and Li et al. (44).
The detection of ALK-rearranged CTCs corresponding to tumor biopsies from the same patients was also presented by Tan et al. (45). In 2015, we reported that rearrangement in the ROS1-tyrosine kinase gene (present in 1% of NSCLC) can be detected in CTCs using FA- FISH (46). We have used FA-FISH combined with fluorescent staining and coupled to the semi-automated microscopy technique. Adapted methodology gave us the possibility to efficiently detect ROS1 rearrangements in CTCs from ROS1-rearranged NSCLC patients.
Following enrichment and identification, the molecular characterization of single CTCs requires technology for their isolation, which can be performed by FACS, DEPArray, microfluidic devices or laser capture microdissection. Several overviews and technology articles of the CTC isolation methods used for the genetic analysis have been already published (47-52).
Except their high heterogeneity, CTCs are also, comparing to the other blood nucleated cells, characterized by increased fragility, which make them even more difficult to isolate. Nowadays many efforts are taking to preserves the viability and enables isolation of intact CTCs with high quality DNA, RNA and proteins. Some promising results have been obtained using EPISPOT method (53), density gradient centrifugation together with telomerase-based assay (54) and recently microfluidic isolation by the CTC-iChip preceded by hypothermic preservation of blood (55). Those approaches might be particularly useful for the characteristic of whole-transcriptome alterations in CTCs. Some laboratories use these methods alone or in combination depending on the type of cancer and the subsequent type of analysis.
Molecular and functional characterization of CTCs
CTCs have the capacity to circulate from one metastasis to another, and may represent cancer genomes issued from multiple metastatic sites (56,57). CTCs may therefore inform on tumor heterogeneity and underlying oncogenic alterations which could serve as predictors of drug sensitivity or resistance. Molecular characterization of CTCs may occur predictive for therapy selection (58).
Our group showed that tumor heterogeneity, evaluated by FA-FISH detection of ROS1 gene copy number, was significantly higher in CTCs at the baseline comparing with corresponding tumor biopsies in the three patients that presented either disease stability or partial response to treatment. Progression during crizotinib treatment observed in the two patients increased significantly the copy number in ROS1-rearranged CTCs. We show that non-invasive blood sample containing CTCs might serve for detection of ROS1 rearrangement in NSCLC patients, as mentioned above. Furthermore, our results show that the CTCs from ROS1-rearranged NSCLC patients display substantial heterogeneity of ROS1-gene aberrations accompanied by a high level of chromosomal instability (CIN). Although technically challenging, the experimental process to detect predictive biomarkers such as rearrangements of ALK, ROS1, RET genes in CTCs form NSCLC has been established (42,43,46,59). Even though still more collaborative research is needed to transfer research improvement into the clinic, these studies provide new perspectives for identifying NSCLC patients eligible for TKI therapy. Non-invasive serial molecular analyses of CTCs performed at different time-points upon therapy may potentially be of great utility to inform therapeutic decisions in NSCLC patients treated by sequential TKI.
To fully understand discrepancies between immunohistochemical analyses (IHC) compared with FISH in predicting response and outcome to ALK inhibitors, further evaluation of several biological features such as the variation of the native ALK copy number as well as the c-MET and ROS1 protein expression could be considered (60,61). The utility of CTCs for MET expression assessment in NSCLC patients has been confirmed (62). Additionally, CTCs has been employed in order to assess the expression of predictive biomarkers for immunotherapy, which is of particular interest for NSCLC patients, since the new clinical trials targeting immune checkpoints are ongoing. CTCs and corresponding tumor biopsies of patients with advanced NSCLC were evaluated for PD-L1 expression. Patients that present high expression of PD-L1 on CTCs, even though their tumor biopsies are negative for PD-L1, they could potentially benefit from immunotherapy (63,64). Parallel evaluation of plasma biomarkers such as different cytokines (e.g., IL-8) could also inform about anti-PD1/PD-L1 treatment efficiency (65).
Overall mentioned studies on CTCs concerned their prognostic and pharmacodynamic significance, identification of predictive biomarkers and utility in the real-time monitoring of disease (43,46,59,61,63). Even though that CTCs are considered as the metastasis seeding elements, only a minority of CTCs with the cancer stem cell properties and metastatic capability retain all the necessary features for survive in the circulation, extravasate and finally form metastases (66). Thus, a key objective right now is the genetic and epigenetic profiling of CTCs and transferring these cells to mouse models to investigate their functional capability, signaling pathways alterations and following drug-screening assays (Figure 2). In metastatic breast cancer, Baccelli et al. reported first time a subpopulation of CTCs with a tumor initiating CD45-EpCAM+CD44+CD47+cMet+ phenotype (24). Up to now very few groups, managed to established functional CTC-derived xenograft (CDX) models. The CDX derived from CTC of SCLC patient has been shown to mimic the donor patient’s response to chemotherapy (67), which validate these kinds of models in the pharmacologic study. The first report showing the tumorigenicity of CTC issued from metastatic patients with NSCLC was published by Morrow et al. (68). Molecular analysis of the CDX confirmed its tumor origin and RNA and protein profiling of CDX showed its mesenchymal phenotype, indicating the functional importance of mesenchymal CTCs (68). The CDX models have been also derived from the melanoma patients (69). CDX-bearing mice developed metastases, which were confirmed to have the human melanoma origin. Our team recently established CDX models derived from CTC of prostate cancer and NSCLC patients (70,71). Additionally research showed the possibility to expand CTCs from early stage lung cancer in a three dimensional co-cultures (72). Extremely low CTC concentration in blood and complicated process of their isolation, together with the limited knowledge about their functional properties, makes the generation and maintenance of CTC in vitro culture remarkably rare. The first CTC cell line has been established from colorectal cancer patient (73). This cell line displayed the phenotypic characteristic of patient’s tumor and tumorigenic potential after engraftment to immunodeficient mice. Intensive advances in the establishment of novel in vitro and in vivo models from CTCs are remarkably increasing the knowledge about the CTC’s biology and open the opportunity to multiple drug testing. Next to single cell genomic analysis the CTC derived cell lines and CDX would indeed allow a full characterization of molecular mechanisms leading to evolution of tumor disease.
Perspectives and advances in research to characterize the origins of CTCs’ heterogeneity
Genetic intratumor heterogeneity fostered by genome instability has a central role in cancer evolution. Indeed, chromosome instability was related with an increased risk of relapse or death of NSCLC patients, and the potential value of chromosome instability as a prognostic predictor has been proposed (74). Some reports show that CIN may provide a source of metastatic potential (75-77). Particularly chromosomal doubling caused by missegregation of chromosomes and cytokinesis failure has been shown to accelerate cancer evolution (78). By analyzing multiple spatially separated tumor and metastasis biopsies increased amount of subclonal copy number alterations (CNA) were associated with disease relapse and death in NSCLC patients. The importance of chromosome instability (CIN) in acquiring resistance has been shown in ALK-rearranged tumors. Our group evaluated by FA-FISH the ALK-rearranged CTC from NSCLC patients under the crizotinib therapy and we correlated the different ALK-FISH patterns with the evolution on crizotinib treatment (59). CTCs were divided into different subgroups according to the existence of ALK-rearrangements and/or ALK copy number gain (ALK-CNG). In line with the earlier report on tumor biopsies, at the baseline of crizotinib treatment no significant report was observed between the amount of ALK-rearranged or ALK-CNG CTCs and PFS. However, we showed that the diminution of the amount of CTCs presenting ALK-CNG upon crizotinib treatment was positively related to longer PFS (59). This research showed that successive analysis of CTCs by FA-FISH may deliver a predictive biomarker for therapy outcome in NSCLC and may stratify patients that present the increased risk of early resistance (59,60). Another example of a predictive role of CIN characterized in CTC has been described for SCLC patients. The minimally invasive CTC-profiling of CNA gave promised results in the classification of chemosensitive versus chemorefractory SCLC patients (79). Even though additional researches are needed to elucidate the clinical utility of CIN profiling in CTCs, these works present a proof-of-concept for the role of genomic instability in the therapy outcome choice. The widespread tumor heterogeneity largely driven by genome doubling and chromosomal instability has been recently described based on the multiple biopsies from lung tumors (80). The important interest relies more often nowadays on liquid biopsies, which not only would be useful for identifying predictive biomarkers of sensitivity or resistance but also increased our knowledge about the heterogeneity driven by genomic instability of CTCs. Recent report presented the phylogenetic ctDNA profiling to characterize the subclonal dynamics of relapsing NSCLC and predict chemotherapy resistance (81) validating at the same time the importance of using liquid biopsies to follow subclonality of tumor development. Rapidly advancing technology would enable to characterize the subclonal evolution of metastasis-initiating CTCs considering in particular their single cell genomic instability and heterogeneity.
Summary conclusions
Liquid biopsy in a single non-invasive blood test may deliver valuable source of biomarkers in NSCLC. The presence of CTCs is associated to worse prognosis and the risk of early recurrence in numerous clinical trials of different cancers. Examination of CTCs at the molecular level carries a great potential to decipher the biology of cancer dissemination and inform on potential targets and signaling networks relevant to therapeutic interventions. Shedding the light to the mechanism of tumor heterogeneity in CTCs would allow for a better understanding of cancer progression with potential improvement of therapeutic strategies to overcome resistance to TKI. Additionally, monitoring of acquired resistance mutations in CTCs may allow adapting sequential therapy with TKI. The potential to interrogate the tumor genome sequentially and systematically, by incorporating CTCs, ctDNA and miRNA analysis into clinical trials is highly recommended to provide unique insights into tumor heterogeneity, evolution and emergence of resistance to treatment from the multiple sites.
Acknowledgements
The authors are grateful to the patients and their families.
Funding: E Pailler was supported by the Fondation pour la Recherche Médicale (grant No. FDT20150532072). V Faugeroux was supported by the Fondation pour la Recherche Médicale (grants No. FDT20160435543). The authors are grateful for the research support of the Fondation de France (grant No. 201300038317), the Fondation ARC pour la Recherche sur le Cancer (grant No. 20131200417), Innovative Medicines Initiative 11th Call CANCER ID (IMI-JU-11-2013, 115749), Institut National du Cancer (PRT-K14-032) and Agence Nationale de la Recherche (ANR-CE17-0006-01).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Shames DS, Wistuba II. The evolving genomic classification of lung cancer. J Pathol 2014;232:121-33. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Camidge DR, Pao W, Sequist LV, et al. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. [Crossref] [PubMed]
- Nguyen KS, Kobayashi S, Costa DB, et al. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer 2009;10:281-9. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small- cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK- positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-19. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK- positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK- positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Perez CA, Velez M, Raez LE, et al. Overcoming the resistance to crizotinib in patients with non-small cell lung cancer harboring EML4/ALK translocation. Lung Cancer 2014;84:110-15. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Katayama R, Friboulet L, Koike S, et al. Two novel ALK mutations mediate acquired resistance to the next generation ALK inhibitor alectinib. Clin Cancer Res 2014;20:5686-96. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol 2013;31:1105-11. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Solomon B, Shaw A, Ou S, et al. OA 05.06 phase 2 study of lorlatinib in patients with advanced ALK+/ROS1+ non-small-cell lung cancer. J Thorac Oncol 2017;12:S1756. [Crossref]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Baccelli I, Schneeweiss A, Riethdorf S, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539-44. [Crossref] [PubMed]
- Ashworth TR. A Case of Cancer in Which Cells Similar to Those in the Tumors Were Seen in the Blood after Death. Australas Med J 1869;14:146-9.
- Levy B, Hu ZI, Cordova KN, et al. Clinical Utility of Liquid Diagnostic Platforms in Non-Small Cell Lung Cancer. Oncologist 2016;21:1121-30. [Crossref] [PubMed]
- Sestini S, Boeri M, Marchiano A, et al. Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget 2015;6:32868-77. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Hofman V, Ilie M, Long E, et al. Detection of circulating tumor cells from lung cancer patients in the era of targeted therapy: promises, drawbacks and pitfalls. Curr Mol Med 2014;14:440-56. [Crossref] [PubMed]
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Hirose T, Murata Y, Oki Y, et al. Relationship of circulating tumor cells to the effectiveness of cytotoxic chemotherapy in patients with metastatic non-small-cell lung cancer. Oncol Res 2012;20:131-7. [Crossref] [PubMed]
- Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel) 2014;6:153-65. [Crossref] [PubMed]
- Juan O, Vidal J, Gisbert R, et al. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin Transl Oncol 2014;16:637-43. [Crossref] [PubMed]
- Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer 2011;105:847-53. [Crossref] [PubMed]
- Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non- small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. [Crossref] [PubMed]
- Lindsay CR, Faugeroux V, Michiels S, et al. A prospective examination of circulating tumor cell profiles in non-small-cell lung cancer molecular subgroups. Ann Oncol 2017;28:1523-31. [Crossref] [PubMed]
- Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Provencio M, Pérez-Callejo D, Torrente M, et al. Concordance between circulating tumor cells and clinical status during follow-up in anaplastic lymphoma kinase (ALK) non-small-cell lung cancer patients. Oncotarget 2017;8:59408-16. [Crossref] [PubMed]
- Li W, Zhang Z, Guo L, et al. Assessment of cytology based molecular analysis to guide targeted therapy in advanced non-small-cell lung cancer. Oncotarget 2016;7:8332-40. [PubMed]
- Tan CL, Lim TH, Lim TK, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [PubMed]
- Pailler E, Auger N, Lindsay CR, et al. High Level of Chromosomal Instability in Circulating Tumor Cells of ROS1-Rearranged Non-Small-Cell Lung Cancer. Ann Oncol 2015;26:1408-15. [Crossref] [PubMed]
- Ross K, Pailler E, Faugeroux V, et al. The potential diagnostic power of circulating tumor cell analysis for non-small-cell lung cancer. Expert Rev Mol Diagn 2015;15:1605-29. [Crossref] [PubMed]
- Swennenhuis JF, Terstappen L. Sample Preparation Methods Following CellSearch Approach Compatible of Single-Cell Whole-Genome Amplification: An Overview. Methods Mol Biol 2015;1347:57-67. [Crossref] [PubMed]
- Mesquita B, Rothwell DG, Burt DJ, et al. Molecular analysis of single circulating tumour cells following long-term storage of clinical samples. Mol Oncol 2017;11:1687-97. [Crossref] [PubMed]
- Neumann MH, Schneck H, Decker Y, et al. Isolation and characterization of circulating tumor cells using a novel workflow combining the CellSearch® system and the CellCelector Biotechnol Prog 2017;33:125-32. [Crossref] [PubMed]
- Premasekharan G, Gilbert E, Okimoto RA, et al. An improved CTC isolation scheme for pairing with downstream genomics: Demonstrating clinical utility in metastatic prostate, lung and pancreatic cancer. Cancer Lett 2016;380:144-52. [Crossref] [PubMed]
- Swennenhuis JF, Reumers J, Thys K, et al. Efficiency of whole genome amplification of single circulating tumor cells enriched by CellSearch and sorted by FACS. Genome Med 2013;5:106. [Crossref] [PubMed]
- Ramirez JM, Fehm T, Orsini M, et al. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin Chem 2014;60:214-21. [Crossref] [PubMed]
- Dorsey JF, Kao GD, MacArthur KM, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non-small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: pilot study results. Cancer 2015;121:139-49. [Crossref] [PubMed]
- Wong VC, Ko JM, Lam CT, et al. Succinct workflows for circulating tumor cells after enrichment: From systematic counting to mutational profiling. PLoS One 2017;12:e0177276. [Crossref] [PubMed]
- Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353-7. [Crossref] [PubMed]
- Hong MK, Macintyre G, Wedge DC, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 2015;6:6605. [Crossref] [PubMed]
- Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells- biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. [Crossref] [PubMed]
- Pailler E, Oulhen M, Borget I, et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non-Small Cell Lung Cancer Patients. Cancer Res 2017;77:2222-30. [Crossref] [PubMed]
- Ilie MI, Bence C, Hofman V, et al. Discrepancies between FISH and immunohistochemistry for assessment of the ALK status are associated with ALK 'borderline'-positive rearrangements or a high copy number: a potential major issue for anti- ALK therapeutic strategies. Ann Oncol 2015;26:238-44. [Crossref] [PubMed]
- Ilie M, Long-Mira E, Bence C, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol 2016;27:147-53. [Crossref] [PubMed]
- Ilie M, Szafer-Glusman E, Hofman V, et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced-stage lung cancer patients. Oncotarget 2017;8:26112-21. [Crossref] [PubMed]
- Ilie M, Hofman V, Dietel M, et al. Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch 2016;468:511-25. [Crossref] [PubMed]
- Nicolazzo C, Raimondi C, Mancini M, et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep 2016;6:31726. [Crossref] [PubMed]
- Hofman P. Liquid biopsy as a potential tool for using predictive biomarkers in immunotherapy ISMRC 2018. Available online: http://www.ismrc2018.com/upload/programme/programme.pdf
- Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016. Cell 2016;166:21-45. [Crossref] [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Morrow CJ, Trapani F, Metcalf RL, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol 2016;27:1155-60. [Crossref] [PubMed]
- Girotti MR, Gremel G, Lee R, et al. Application of Sequencing, Liquid Biopsies, and Patient- Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discov 2016;6:286-99. [Crossref] [PubMed]
- Faugeroux V, Pailler E, Deas O, et al. Establishment and characterization of a unique circulating tumor cells derived xenograft (CDX) in prostate cancer. Clin Res 2018;78:abstr 5600.
- Faugeroux V, Pailler E, Deas O, et al. Development and characterization of novel non-small-cell lung cancer (NSCLC) circulating tumor cells derived xenograft (CDX) models. Clin Res 2018;78:abstr 2597.
- Zhang Z, Shiratsuchi H, Lin J, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014;5:12383-97. [PubMed]
- Cayrefourcq L, Mazard T, Joosse S, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res 2015;75:892-901. [Crossref] [PubMed]
- Jamal-Hanjani M, Gareth AW, McGranahan N, et al. Tracking the evolution of Non-small- cell lung cancers. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Dewhurst SM, McGranahan N, Burrell RA, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014;4:175-85. [Crossref] [PubMed]
- Fujiwara T, Bandi M, Nitta M, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005;437:1043-7. [Crossref] [PubMed]
- Maley CC, Galipeau PC, Finley JC, et al. Tetraploidy and chromosomal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 2006;38:468-73. [Crossref] [PubMed]
- Bakhoum SF, Ngo B, Laughney AM, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467-72. [Crossref] [PubMed]
- Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]