
Noninvasive biomarkers for lung cancer diagnosis, where do we stand?
Introduction
Most lung cancer diagnosis are preceded by detection of a nodule by chest computed tomography (CT) imaging. The majority of those are still found incidentally. The epidemic is however growing given the implementation of lung cancer screening. Screening-detected nodules are on the rise, with an estimated 1.5 million nodules detected per year over an estimated 5 million people (1). Several landmark studies in the 2010’s demonstrated that early detection of lung cancer is a powerful approach to reducing mortality and improving patient outcomes in the deadliest cancer worldwide. Lung cancer is often asymptomatic until later stage. Screening of high-risk individuals allows detection of lung nodules that are or may become cancerous at a much earlier timepoint (2). The National Lung Screening Trial demonstrated that low-dose CT screening of individuals with known risk factors has a 20% relative reduction in lung-cancer mortality when compared to X-ray screening (3). Similar results were observed in the Dutch-Belgian Randomized Lung Cancer Screening Trial (NELSON) (4), with a 26% reduction in 10-year cancer mortality among men when screened by CT compared to no screening, and the Multicentric Italian Lung Detection (MILD) trial resulted in 39% lower lung cancer mortality at 10 years (5). Other screening trials have demonstrated similar effects, including the Italian lung study (ITALUNG) (6), Detection and screening of early lung cancer with novel imaging technology (DANTE) (7), Danish lung cancer screening trial (DLCST) (8), the German lung cancer screening intervention trial (LUSI) (9), and the UK lung cancer screening (UKLS) (10). The conclusion is in: Low dose chest CT screening of high-risk individuals reduces lung cancer mortality.
However, with this success comes new challenges. Currently, an estimated 1.5 million new lung nodules are detected annually in the United States alone, and as screening programs are more widely implemented, this number will continue to rise (1). Lung nodules detected on screening CT scans exhibit a false positive rate greater than 95%. Lung nodules that are calcified or very small are at a very small risk of lung cancer, but most nodules detected through screening or incidentally require follow-up diagnostic procedures. The options to obtain further diagnostic information include bronchoscopy, fine needle aspiration, transthoracic needle aspiration, or surgical biopsy (11). The alternative to tissue-based assessment includes sequential CT to assess growth, FDG-PET and contrast CT to further assess risk of malignancy.
For many of the cancers with widespread screening [breast (12), colorectal (13), cervical, prostate, and skin], a positive screening result can be quickly followed up with a tissue biopsy at minimal extra risk to the patient (14). This is not the case in lung cancer. Invasive procedures often require general anesthesia and its attendant risks, including significant rates of pneumothorax (15,16). A recent cost-benefit analysis showed that among Medicare claims over 40% of the total cost in management of lung cancer was attributed to benign patients with an invasive procedure (17).
Diagnostic biomarkers in clinical practice
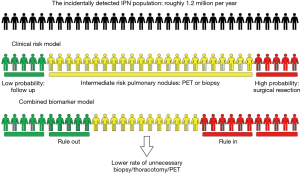
Because invasive approaches are associated with morbidity, cost and delay in diagnosis, the rationale for the development of non-invasive strategies is very strong. Like we experience in cardiovascular (troponin) (18), endocrine (HbA1C) or infectious (HIV viral load) diseases evaluation, diagnostic biomarkers have brought tremendous benefits to clinical practice. Most lung cancer diagnoses are preceded by a positive chest CT. A biomarker to rule in or out lung cancer among patients with indeterminate pulmonary nodules (IPNs) would have tremendous clinical benefit in reducing the rate of benign thoracotomies, the rate of invasive procedures, the time to diagnosis and cost among important outcomes, Figure 1. A successful biomarker would modify the management of lung nodules and lead to improvement of these outcomes by providing actionable information. This review does not comprehensively review all circulating biomarkers, but rather highlights a few studies that have attempted to address the non-invasive diagnosis of lung cancer among individuals presenting with IPNs with a focus on circulating biomarkers.
Blood protein biomarkers
Blood biomarkers represent one of the most attractive methods for diagnostic evaluation due to their low risk to the patient and ease of access. The two most studied family of blood biomarkers for diagnostics are proteins and micro-RNA panels. Many signatures have been studied, so in this review we choose to focus on studies that have implemented a blood biomarker for the purpose of distinguishing benign from malignant pulmonary nodules. Here we have chosen to focus on studies occurring in the years 2010–2019.
Table 1 contains a summary of studies which have implemented molecular blood biomarkers, and more specifically, oligonucleic biomarkers as a diagnostic test in nodule cohorts. Here we have drawn attention to the biomarker panels, the population size in the training and, if applicable, validation cohorts broken down by the number of patients with a positive cancer diagnosis, patients with a nodule determined to be noncancerous, and patients with no nodule. The composition of no-nodule populations varies from study-to-study, as some studies include healthy individuals in the “no nodule” category as control and some studies include patients with benign lung disease. We have also listed the performance of the biomarker test in terms of reported sensitivity/specificity. It should be kept in mind that comparing the sensitivity and specificity is not the ideal way to compare multiple biomarker tests when the tests were trained and validated on different populations. Additionally, a single sensitivity/specificity reported for a dichotomous test is not indicative of the true performance of the continuous variable resulting from these biomarker tests. However, when summarizing these results, we feel that reporting the sensitivity/specificity, as reported by the authors of these studies, serves to showcase how the authors intended their biomarker test to be utilized. For example, a study with a moderate sensitivity and an outstanding specificity can be utilized as a definitive rule-in test, and conversely an outstanding sensitivity and mediocre specificity can be a helpful rule-out test. For these reasons, we caution against using the listed sens/spec values in isolation as an indication as to which test performance is “better”.
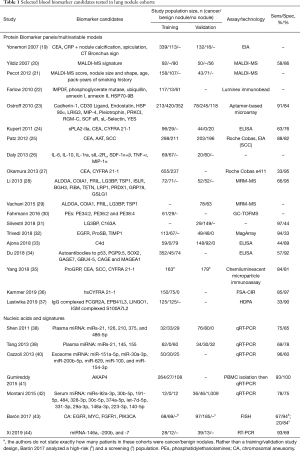
Full table
Prior to 2010, several diagnostic biomarker studies attempting to discriminate benign from malignant IPNs were published. While not meeting all criteria we defined above, one is worth mentioning for their contribution to the field. Yonemori 2007 used a model incorporating serum C-reactive protein (CRP) and carcinoembryonic antigen (CEA) in combination with the presence or absence of nodule spiculation, calcification, and CT bronchus sign in a population of 452 patients (113 benign nodules and 339 malignant) (19). Their model was compared against an expert chest radiologist making predictions. In a validation set, the experienced chest radiologist outperformed the model, with an AUC of 0.905 vs. 0.858. The two antigens, CRP and CEA, have repeatedly been demonstrated to have diagnostic potential for lung nodules and are used in several of the subsequent studies (45,46). CRP is associated with inflammation. CEA is an umbrella term for a family of closely related glycoproteins that aide in cellular adhesion. CEA typically is downregulated prior to birth, but it is hypothesized to be upregulated in several epithelial cancers to contribute to metastasis. Kupert et al. demonstrated that CEA was useful in combination with secretory phospholipase A2-IIa, a phospholipid hydrolase enzyme that mediates the release of several precursors to eicosanoids, which regulate inflammation, immunity, and tumorigenesis (24). sPA2-IIa, which had previously been demonstrated to be elevated in prostate cancer patients as well, perfectly discriminated between 96 lung cancers and 20 healthy donors, but the discrimination between lung cancer and 29 benign nodules was less accurate (AUC of 0.68). Also included in this study was CYFRA 21-1, a fragment of cytokeratin 19 which is released from epithelial cells upon cell death. CYFRA 21-1 has been used as a marker in other epithelial cancers and as a marker of epithelial inflammation. The three-protein panel of sPA2-IIa, CEA, and CYFRA 21-1 quantified by ELISA was shown to outperform sPA2-IIa alone. Interestingly none of these biomarkers are cancer-specific, but rather markers of inflammation, which is a common problem across cancer biomarker research. Okamura et al. assessed the level of CEA and CYFRA 21-1, and showed improved diagnostic specificity in a cohort of 655 lung cancer patients and 237 patients with benign lung disease when combining the two biomarkers (27).
CEA was again investigated by Patz et al., this time in the context of a nodule cohort of 298 cancers and 211 benign nodules, in combination with α1-antitrypsin (AAT) and squamous cell carcinoma (SCC) antigen. Alpha 1 antitrypsin, also known as serum trypsin inhibitor, is an inflammatory marker used in T-cell migration, and SCC is a protease inhibitor found in squamous epithelium that have been shown to promote tumor growth. Malignancy was associated with increased levels of CEA, AAT, and SCC. In this case, a validation cohort (n=203 cancers and 196 benign nodules) demonstrated similar performance, with both cohorts reporting sensitivity/specificity above 80%. Again, CEA, CYFRA 21-1, SCC, and ProGRP measured by chemiluminescent microparticle immunoassays on the ARCHITECT platform were used in large cohort study in China and demonstrated improved performance over standard ACCP risk assessment (35).
Our own work has focused on implementation of the free solution assay (FSA) method measured by the compensated interferometric reader (CIR) for high sensitivity biomarker quantification. Like others, we have investigated CYFRA 21-1, but by focusing on increased biomarker sensitivity, we have improved the potential utility of the biomarker. The FSA method capitalizes on changes in solution dielectric constant when a probe molecule binds to a target molecule to quantify the amount of bound target (47). By adding an excess quantity of a probe molecule, typically an antibody, aptamer (48), or known inhibitor, to a patient serum, plasma, or urine sample (49), and a non-binding control to a matched aliquot of the same sample, highly specific binding can be quantified while minimizing background signals. The method capitalizes on a self-referencing technique where the patient’s serum or plasma sample is split into two aliquots of equal volume, and the probe (a monoclonal antibody) is added to one aliquot while a RI-matched solution is added to the other, and the difference in signal is measured between the two. In a case control study of 150 cancers and 75 benign nodules, the high sensitivity CYFRA 21-1 assay demonstrated a sensitivity/specificity of 85%/97%, which outperformed a standard electrochemiluminescence ELISA measurement of CYFRA 21-1 on the same cohort. In this study, lower limits of quantification enabled more accurate measurement of CYFRA 21-1 levels at low concentrations when using hsCYFRA 21-1, and as a result, many controls were measured to have much lower biomarker levels, increasing the discrimination from cases (36).
Yildiz et al. 2007 presented a matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) signature that could distinguish lung cancer from benign lung disease (20). The signature resulted from an analysis of serum peptides, but interestingly, the signature was derived and tested without a full determination of the underlying peptides responsible for the signature. In 2012, Pecot et al. demonstrated validation of this signature and showed added value to imaging and clinical variables for diagnosis when combining the MALDI signature with CT imaging variables (nodule size and shape) and clinical variables (age and smoking pack years) in a population of patients with indeterminate nodules (21). The added value of the MALDI signature was only significant in a subpopulation of 5–20 mm nodules, however this is the most clinically challenging subpopulation.
In 2013, Li et al. used multiple reaction monitoring (MRM) MS to develop a proteomic classifier that assessed 13 proteins. Six of these proteins were secreted (ISLR, BGH3, FIBA, TSP1, TETN, COIA1), two were membrane receptors (LG38P, LRP1), and 5 were cytoplasmic (FRIL, PRDX1, GRP78, ALDOA, GSLG1) (28). The classifier was trained in a cohort of 72/71 cancers/nodules and then validated in a second cohort of 52/52 and exhibiting high sensitivity and lower specificity (83%/29% for an assumed prevalence of 20%). This study was then validated in a clinical utility study in 2015 by the same group, using a refined panel with five proteins (ALDOA, COIA1, FRIL, LG3BP, TSP1), and achieving similar results in a cohort of 78 cancers and 63 benign nodules (29). Use of this classifier would have resulted in a 32% reduction in unnecessary surgeries and a 31.8% in total invasive procedures, while suggesting that 18% of malignant nodules would go for surveillance CT, which is favorable to a multisite observational study which puts this number at 24.5% in current pulmonary practice.
This study was followed-up with the Pulmonary Nodule Plasma Proteomic Classifier (PANOPTIC) trial (Clinicaltrials.gov trial No. NCT01752114) (31). The prospective observational trial of 685 patients with 8–30 mm lung nodules used a clinical risk prediction that incorporated the relative abundance of two plasma proteins, LG3BP and C163A to identify likely benign nodules (50). The integrated classifier outperformed PET, validated lung nodule risk models, and physician probability estimates. This trial focused on ruling out patients with a pre-test probability of cancer of <50%, and the preliminary analysis of clinical utility demonstrated that 40% fewer procedures would be performed on benign nodules within this population. While the true clinical utility of the biomarker is yet to be tested, these results are promising for the inclusion of protein biomarkers in risk stratification (51).
Another mass-spec based study analyzed the levels of serum phosphatidylethanolamines (PEs). Untargeted GC-TOFMS and HILIC-qTOF-MS based metabolomics was used to determine three PEs with the best accuracy in a cohort of 62 malignant nodules and 29 benign nodules (30). Further validation of PEs as biomarkers should be enlightening.
An autoantibody-based test based upon the Luminex Immunobead platform was published by Farlow et al. in 2010 (22). This panel incorporated several proteins that were found to be highly different between cancers and controls (13 benign nodules, 30 COPD patients, and 31 healthy controls) by 2D western blot followed by MS identification of promising markers. The signature included: (I) Inosine-5’-monophosphate dehydrogenase (IMPDF), an important catalyst of oxidation of inosine monophosphate to xanthosine monophosphate, a rate limited step in synthesis of guanine nucleotides. (II) Phosphoglycerate mutase, a glycolysis enzyme. (III) Ubiquillin, a ubiquitin like protein thought to functionally link the ubiquitination machinery to the proteasome to affect in vivo protein degradation. (IV) Annexin I and (V) annexin II, which are lipocortins that are involved in the inhibition of inflammation by glucocorticoids. (V) HSP70-9B, a member of the heat shock protein 70 family thought to be involved in the control of cellular proliferation. This 6-marker panel was used in a “Cart” algorithm to classify patients and demonstrated a low false positive rate (4%).
Daly et al. demonstrated a similar performance using a panel of 7 circulating biomarkers including IL-6, IL-10, IL-1ra, sIL-2Rα, and stromal cell-derived factor-1α+β . The added performance of ILs in this panel is not unexpected, as they, like many other biomarkers in this field, are integral in the immune response. This panel achieved a perfect sensitivity and negative predictive value (100%) with a specificity of 52% (26).
Autoantibody tests have the promise of high specificity but relatively low sensitivity for the low prevalence of common autoantibodies identified in lung cancer. Tumor-associated antigens targeted included CAGE, GBU 4–5, p53, MAGE A4, HuD, NY-ES0-1 and SOX-2. This is an autoantibody test in the plasma of high-risk individuals. We recently tested its clinical significance in a registry study where a total of 451 patients (32%) had at least one nodule, leading to 296 eligible patients after exclusions, with a lung cancer prevalence of 25%. In 4- to 20-mm nodules, a positive test result represented a greater than two-fold increased relative risk for development of lung cancer as compared with a negative test result. Also, when the “both-positive rule” for combining binary tests was used, adding EarlyCDT-Lung to risk models improved diagnostic performance with high specificity (>92%) and positive predictive value (>70%) (52,53). This classifier was designed to have high PPV, achieving >25% as required for clinical practice. Such a biomarker may be worth proposing as a surveillance program for high-risk individuals with lung nodules, if the classifier passes a probability threshold that avoids futile further clinical work-up or a missed chance for cure.
A blood based proteomic profile was developed in 2018 by Trivedi et al. which utilized a novel immunoassay sensing technique, MagArray, to detect 3 circulating proteins, EGFR, ProSB, and TIMP1. The MagArray platform uses a sandwich strategy with a magnetic nanoparticle conjugated to the detection antibody. Changes in the magnetic field at the sensor surface correlate with antigen concentration (54). The initial training cohort consisted of 113 cancerous and 67 benign nodules, with a validation of 49 cancer/48 benign nodules, demonstrating a sensitivity of 94% (32). Subsequent work by the same group expanded the panel to also include CEA and NAP2, and demonstrated a sensitivity/specificity of 76%/82% in a testing cohort of 144 patients (55).
Fragments of complement component 4 (C4), specifically fragments containing the C4D moiety, were investigated as a circulating marker of malignancy by Ajona et al. in 2018. In contrast to the group’s previous studies, specific C4D outperformed total C4 derived fragments in a study comparing 39 cancers with 39 age and smoking matched controls. Measurement in plasma outperformed bronchoalveolar lavage. Assessment performance of C4D in a nodule cohort demonstrated improved accuracy over CT, with especially strong specificity.
High density protein arrays (HDPA) represent a promising approach for biomarker discovery, validation, and use as a large panel (56). This technology uses several thousand (3,000–17,000, depending upon the configuration) different antibodies printed in an array. After a sample, such as human plasma, is introduced, any binding interaction with the immobilized antibodies will then be measured using fluorescence. In this method, many thousands of targets can be assayed simultaneously with the sensitivity of a more involved ELISA immunoassay. An advantage of this technology is the ability to simultaneously measure the protein in its native state, glycosylated through post-translational-modification, and bound to an autoantibody. Any protein that shows a consistent cancer-specific change in two or three of these can be considered a “hybrid marker”. In addition to the extra dimensionality that the hybrid markers provide, it is hypothesized that hybrid markers could enable less variation between different individuals because one of the markers can serve to standardize the measurement (57). A HDPA was recently implemented to profile autoantibodies isolated from B cells in resected lung tumors, and the most significantly elevated autoantibodies were used in a panel for plasma analysis. Thirteen of these B-cell-derived antibodies were selected, and five were found to be significantly higher in plasma from patients with non-small cell lung cancer when compared to plasma from patients with benign nodules (37). A logistic regression was used to build a classifier of four autoantibodies (FCGR2A, EPB41L3, and LINGO1 IgG-complexed autoantibodies and S100A7L2 IgM-complexed autoantibody) that enabled discrimination between cases and controls in a 250-patient cohort (ROC AUC of 0.737). Interestingly, these autoantibodies may provide novel information about the malignant nodules because none of the antibodies were correlated with any of the currently used clinical risk factors (smoking status, age, or nodule volume).
Another study investigating autoantibodies developed a panel of 7 AAbs (to p53, PGP9.5, SOX2, GAGE7, GBU4-5, CAGE and MAGEA1) in a cohort of 305 NSCLCs, 47 SCLCs, 45 benign nodules, and 74 controls (34). When combined with CT, this signature produced a specificity of 91.6% for cancers vs. benign nodules, with a sensitivity of 56.5%.
An alternate approach to lung nodule management is to confirm a benign nodule’s disease state, as has been done with testing for antibodies against histoplasma capsulatum to rule out benign nodules as granulomas of infectious origin (58). Histoplasmosis is a soil fungi endemic to wet regions (59), which produces granulomas that are radiologically at indeterminate risk for cancer. An initial evaluation showed that all patients who were positive for both an elevated IgG and IgM autoantibodies to histoplasmosis capsulatum were benign, thus conferring to the test an excellent negative predictive value for cancer. While this specific test is appropriate in a small subset of the total IPN population, it is very powerful at confirming benign nodule status when used in the correct circumstances.
Genetic and epigenetic biomarkers
Circulating RNAs have been widely studied to develop diagnostic panels, and several studies have been implemented in nodule cohorts, with promising sensitivity and specificity. In Table 1 we have highlighted several studies which have tested plasma (38,39), serum (42), and exosomal (40) miRNA signatures. Plasma signatures have also been combined with CT features for IPN discrimination (44).
Gumireddy et al. demonstrated a method to capture peripheral blood mononuclear cells (PBMCs), then use RT-PCR to detect a messenger RNA, AKAP4, as a biomarker for IPN diagnosis (41). Another approach to PBMC analysis utilized gene expression in a 29-gene signature (60). This signature enabled a 91% sensitivity, 80% specificity in a cohort of 137 NSCLC patients 91 controls, including 41 with benign lung nodules.
Chromosomal aneusomy (CA) was demonstrated to be a potential biomarker in a study of high risk patients (68 cancers and 69 controls) and a screening cohort (97 cancers and 185 controls) (43). CA was assayed using a FISH analysis for a four-target DNA panel encompassing the EGFR and v-MYC avian myelocytomatosis viral oncogene homolog (MYC) genes, and the 5p15 and centromere 6 regions or the fibroblast growth factor 1 gene (FGFR1) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene (PIK3CA). The four-target DNA panel achieved a sensitivity/specificity of 67%/94% in the high-risk population and 20%/84% in the screening cohort.
Cancer specific methylation of CD01, HOXA9, and TAC1 is a strong biomarker for lung cancer, reported by a study that derived the methylation signature in The Cancer Genome Atlas, and then validated in two independent cohorts of primary samples (61). The methylation signature achieved 100% specificity, with no methylation in 75 TCGA normal samples and seven primary normal samples and achieved between 83% and 99% sensitivity for non-small cell cancer.
Silvestri et al. demonstrated a bronchial genomic signature specifically for nodule patients with nondiagnostic bronchoscopies (62). While this study did not utilize circulating biomarkers, the use of genomic signature on bronchoscopic samples enabled high-sensitivity (96 and 98% across two study cohorts) confirmation of benign status that could avoid further invasive procedures.
Liquid biopsy
Liquid biopsy refers to the analysis of circulating biomarkers from peripheral blood such as circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), methylated DNA, exosomes or tumor-educated platelets (TEPs). These candidate biomarkers offer a new source of cancer-derived products that may reflect the status of the disease. A very recent report demonstrated that integrating CTCs with CEA quantification could aide in diagnosis of IPNs in a cohort of 80 (30 nodules, 50 cancers) (63). Detection of folate receptor (FR)-positive CTCs was achieved using immunomagnetic bead negative enrichment, and FR-positive CTCs were quantitatively detected by ligand-targeted PCR. The combination achieved a sensitivity of 70% and specificity of 79%.
Another novel approach to biomarker use was demonstrated by Cohen et al. which assessed circulating proteins and mutated driver genes in cell-free DNA (CancerSEEK) to diagnose eight cancer types (64). This test would be broadly administered, and a positive test comes with a localization to a specific organ system. The test demonstrated a lung cancer specific sensitivity of 58%, and an excellent specificity of 99%, with only 7 of 812 healthy controls resulting in a positive determination. This approach has not been formally tested in IPNs as of yet. Start a new paragraph here. Very few of these candidate biomarkers have been assessed in a screening setting. Despite implementation of screening guidelines, screening by low dose CT leads to a high number of false positives. Additionally, current screening guidelines only cover a fraction of patients who go on to develop lung cancer (65). For these reasons, investigating circulating biomarkers in the context of screening may prove to be useful in the future.
One such study was by Guida et al., which developed a panel of 4 proteins (CA-125, CEA, Pro-SFTPB, and CYFRA 21-1) in ever smoking patients, provided a sensitivity/specificity of 42%/95% that patients would develop cancer within the next year (66). This proof-of-principle study demonstrated that use of circulating biomarkers have the potential to inform lung cancer risk, and to facilitate screening.
Radiomics
Imaging features are widely used in the non-invasive diagnosis of lung cancer, but in clinical practice they are currently limited to measuring the nodule’s largest diameter and assigning one or more semantic features, such as lobulation, spiculation, density solid/subsolid/part solid and location. However, there is a wealth of information present in the three-dimensional reconstruction of the nodule that has yet to be leveraged in the clinic for more accurate risk assessment. In fact, the most experienced radiologists can look at this wealth of data and make diagnostic predictions that outperform quantitative risk models (67). Despite the accuracy of experienced radiologists, generalizability is difficulty due to the years of training and experience required to obtain this level of expertise (68). Inter-grader variability confounds this challenge to generalizability, even when the radiologists grading the scans are of similar experience level (69,70).
Quantitative structural image analysis incorporates and standardizes more of the imaging data into diagnosis or risk assessment. From the three-dimensional reconstruction of the nodule, many image “features” can be calculated. Some of these relate to the nodule’s size, such as the diameter in in multiple dimensions, or the shape, such as the nodule’s sphericity. Other features can relate to the nodule’s density or heterogeneity, such as Grey-Level-Co-occurrence-Matrix, entropy, or Kurtosis. Several approaches have been developed for this, including analysis of static nodules (71-75) and analyzing the change in quantitative features over time (76).
Another approach for in-depth radiomics analysis, which skips the intermediate step of quantifying nodule parameters, is direct analysis of the CT scans using deep learning. Arteta et al. demonstrated that a convolutional neural network (CNN) trained on CT scans from the NLST outperformed the Brock University model in predicting malignancy for IPNs (77,78). Similarly, Ardila et al. demonstrated that a CNN algorithm could outperform six trained radiologists of different experience levels with absolute reductions of false positives by 11% and false negatives by 5% when analyzing a single scan (79). When multiple scans were available, the CNN’s performance was comparable to the more experienced radiologist. Such an approach to CT analysis could be especially useful in lower-resource settings which do not have access to a team of highly experienced radiologists.
Our own work has focused on developing a Lung Cancer Prediction Convolutional Neural Network (LCP-CNN) model to determine malignancy. The LCP-CNN was trained using CT images of IPNs from the National Lung Screening Trial (NLST), internally validated, and externally tested on cohorts from two academic institutions. The ROC-AUC in the external validation cohorts were 83.5% and 91.9% compared with 78.1% and 81.9 respectively for a commonly used clinical risk model for incidental nodules. Using ACCP rule-in and rule-out guidelines defining low and high-risk categories, the overall net reclassification in the validation cohorts for cancers and benign nodules compared to the Mayo model was 0.34 and 0.30 as a rule-in test, and 0.33 and 0.58 as a rule-out test, for the two validation cohorts, respectfully (80). Similar approaches to automated risk assessment using machine learning or deep learning methods have been reported by several (72,75,81).
A comment on biomarker study design
When conducting biomarker studies or analyzing results from others, researchers should keep in mind that the current clinical standard performs solidly when used in the appropriate context. Clinical guidelines ensure that suspicious nodules are followed, so even if a definitive diagnosis is not reached at the initial time of nodule detection, the nodule can typically be diagnosed within a time frame that minimizes extensive stage shift (82). Because the use of a biomarker test for lung cancer diagnosis (as opposed to risk assessment) will nearly always follow the discovery of an IPN, diagnostic biomarker studies performed outside of the context of IPN setting are unlikely to lead to significant change in clinical decision making.
Thus, the study design and in particular the selection of appropriate controls for biomarker discovery is critical. Because current clinical decision making is driven by the nodule’s appearance on CT image (e.g., size, shape, and margin), even studies with controls containing radiologically IPNs could be deceptively optimistic or pessimistic. A case-control study where there is a stark contrast in nodule size between cases and controls may present biomarker results that discriminate patients with impressive accuracy, but due to the disparate nodule sizes, the clinical standard decision-making process may already perform very well in this cohort. In such a study design, the biomarker may add very little to the overall discriminatory ability of clinicians, and therefore despite impressive results, may not add much of value to the field. Conversely, a case-control cohort study of patients who have similar known risk factors (include nodule characteristics) may demonstrate moderate biomarker performance, but it may yield significant improvement over current clinical standard.
Additionally, the selection of source material for biomarker (e.g., plasma, serum, exosomes, urine, airway specimens, DICOM images) discovery and validation is important in this regard, both in biology and in the patient cohort selection. Tumors are structurally, molecularly and functionally heterogeneous in both space and time, and therefore capturing this heterogeneity in a biomarker strategy is challenging. While single biomarkers are unlikely to perform well, combination of biomarkers of different biochemical properties and eventually from different source has recently been proposed, and biomarkers selected from the circulation may present such opportunity. Some biomarkers are “shed” from living tissues, while others result from cell death. Molecular entities can undergo structural modifications during cell death or after release into circulation. Although not directly tested in the evaluation of IPNs, strategies combining protein and genomic information for example shows promise (64). This offers new technical challenges of testing a variety of moieties with different technologies and the need for integrating those in a predictive diagnostic model.
A correlated concern is the value added of the test to clinical standard of care. A biomarker test will be useful if it adds information about a patient’s disease status that is not already available to the clinician (83), and that may trigger another management decision. If a biomarker test accurately rules in patients that would have already been suggested for surgery based upon the radiological traits of the nodule, the biomarker has not added significantly to the clinician’s diagnostic toolkit and may not add much value to the field (84-86). A clinician does not need a blood test to suggest a tissue biopsy for a 30 mm nodule with spiculated margins in the chest of a 65-year-old with 80 pack years of smoking history. For this reason, biomarkers that adds value to what is already known by imaging alone for example will be much more valuable than biomarkers that yield duplicate information. Thus, to move biomarker testing in a clinical meaningful way often requires derivation and validation of biomarker tests in the same clinical context.
We therefore encourage researchers to include comparison to the relevant current clinical standard of care when presenting biomarker validation results, and simultaneously, we encourage the community to remain especially skeptical of biomarker studies that omit this crucial benchmark when evaluating the true value of any candidate diagnostic biomarker. Improvements in discriminatory power should be presented with the appropriate tests for significance (87), and when appropriate, assessment of risk classification and reclassification (88-90). This observation is not novel (91), yet reports continue to appear in the literature containing otherwise strong candidate biomarker studies that lack proper evaluation of the true value of the test.
In conclusion, biomarker research is rapidly progressing towards a better understanding of the mechanisms underlying the pathophysiology of lung cancer development and towards the non-invasive management of IPNs. Some of the most useful innovations in the management of IPNs include the possibility to detect the presence or absence of imaging features, circulating protein biomarkers, driver gene mutations with the subsequent choice to decide on a tissue diagnosis or non-invasive follow up. The future management of IPNs could include the application of predictive models integrating longitudinal assessment of clinical, molecular and imaging biomarkers. In the near future, we also hope to see personalized molecular screening available to help identify individuals at highest risk for lung cancer in a cost-effective and non-invasive manner.
Acknowledgments
Funding: This work was supported in part by CA152662 and CA186145 to PPM.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Fabien Maldonado and Robert Lentz) for the series “Novel diagnostic techniques for lung cancer” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflict of interests: Both authors have completed the ICMJE uniform disclosure forms (available at http://dx.doi.org/10.21037/jtd-2019-ndt-10). The series “Novel Diagnostic Techniques for Lung Cancer” was commissioned by the editorial office without any funding or sponsorship. MNK reports patents 10627396 and 10261013 issued. PPM has no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gould MK, Tang T, Liu IL, et al. Recent Trends in the Identification of Incidental Pulmonary Nodules. Am J Respir Crit Care Med 2015;192:1208-14. [Crossref] [PubMed]
- Tammemägi MC, Katki HA, Hocking WG, et al. Selection criteria for lung-cancer screening. N Engl J Med 2013;368:728-36. [Crossref] [PubMed]
- Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N Engl J Med 2020;382:503-13. [Crossref] [PubMed]
- Pastorino U, Sverzellati N, Sestini S, et al. Ten-year results of the Multicentric Italian Lung Detection trial demonstrate the safety and efficacy of biennial lung cancer screening. Eur J Cancer 2019;118:142-8. [Crossref] [PubMed]
- Paci E, Puliti D, Lopes Pegna A, et al. Mortality, survival and incidence rates in the ITALUNG randomised lung cancer screening trial. Thorax 2017;72:825-31. [Crossref] [PubMed]
- Infante M, Cavuto S, Lutman FR, et al. Long-term follow-up results of the DANTE trial, a randomized study of lung cancer screening with spiral computed tomography. Am J Respir Crit Care Med 2015;191:1166-75. [Crossref] [PubMed]
- Wille MM, Dirksen A, Ashraf H, et al. Results of the randomized danish lung cancer screening trial with focus on high-risk profiling. Am J Respir Crit Care Med 2016;193:542-51. [Crossref] [PubMed]
- Becker N, Motsch E, Trotter A, et al. Lung cancer mortality reduction by LDCT screening-Results from the randomized German LUSI trial. Int J Cancer 2020;146:1503-13. [Crossref] [PubMed]
- Field JK, Duffy SW, Baldwin DR, et al. UK Lung Cancer RCT Pilot Screening Trial: baseline findings from the screening arm provide evidence for the potential implementation of lung cancer screening. Thorax 2016;71:161-70. [Crossref] [PubMed]
- MacMahon H, Naidich D, Goo J, et al. Guidelines for management of incidental pulmonary nodules detected on CT Images: from the Fleischner society 2017. Radiology 2017;284:228-43. [Crossref] [PubMed]
- Welch HG, Prorok PC, O'Malley AJ, et al. Breast-cancer tumor size, overdiagnosis, and mammography screening effectiveness. N Engl J Med 2016;375:1438-47. [Crossref] [PubMed]
- Bibbins-Domingo K, Grossman DC, Curry SJ, et al. Screening for colorectal cancer: US preventive services task force recommendation statement. JAMA 2016;315:2564-75. [Crossref] [PubMed]
- Huang H, Wang W, Lin T, et al. Comparison of the complications of traditional 12 cores transrectal prostate biopsy with image fusion guided transperineal prostate biopsy. BMC Urol 2016;16:68. [Crossref] [PubMed]
- Shiekh Y, Haseeb WA, Feroz I, et al. Evaluation of various patient-, lesion-, and procedure-related factors on the occurrence of pneumothorax as a complication of CT-guided percutaneous transthoracic needle biopsy. Pol J Radiol 2019;84:e73-9. [Crossref] [PubMed]
- Boskovic T, Stanic J, Pena-Karan S, et al. Pneumothorax after transthoracic needle biopsy of lung lesions under CT guidance. J Thorac Dis 2014;6 Suppl 1:S99-107. [PubMed]
- Lokhandwala T, Bittoni MA, Dann RA, et al. Costs of Diagnostic Assessment for Lung Cancer: A Medicare Claims Analysis. Clin Lung Cancer 2017;18:e27-34. [Crossref] [PubMed]
- Saenger AK, Beyrau R, Braun S, et al. Multicenter analytical evaluation of a high-sensitivity troponin T assay. Clin Chim Acta 2011;412:748-54. [Crossref] [PubMed]
- Yonemori K, Tateishi U, Uno H, et al. Development and validation of diagnostic prediction model for solitary pulmonary nodules. Respirology 2007;12:856-62. [Crossref] [PubMed]
- Yildiz PB, Shyr Y, Rahman JS, et al. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol 2007;2:893-901. [Crossref] [PubMed]
- Pecot CV, Li M, Zhang XJ, et al. Added value of a serum proteomic signature in the diagnostic evaluation of lung nodules. Cancer Epidemiol Biomarkers Prev 2012;21:786-92. [Crossref] [PubMed]
- Farlow EC, Patel K, Basu S, et al. Development of a multiplexed tumor-associated autoantibody-based blood test for the detection of non-small cell lung cancer. Clin Cancer Res 2010;16:3452-62. [Crossref] [PubMed]
- Ostroff RM, Bigbee WL, Franklin W, et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One 2010;5:e15003. [Crossref] [PubMed]
- Kupert E, Anderson M, Liu Y, et al. Plasma secretory phospholipase A2-IIa as a potential biomarker for lung cancer in patients with solitary pulmonary nodules. BMC Cancer 2011;11:513. [Crossref] [PubMed]
- Patz EF Jr, Campa MJ, Gottlin EB, et al. Biomarkers to help guide management of patients with pulmonary nodules. Am J Respir Crit Care Med 2013;188:461-5. [Crossref] [PubMed]
- Daly S, Rinewalt D, Fhied C, et al. Development and validation of a plasma biomarker panel for discerning clinical significance of indeterminate pulmonary nodules. J Thorac Oncol 2013;8:31-6. [Crossref] [PubMed]
- Okamura K, Takayama K, Izumi M, et al. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer 2013;80:45-9. [Crossref] [PubMed]
- Li XJ, Hayward C, Fong PY, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med 2013;5:207ra142. [Crossref] [PubMed]
- Vachani A, Hammoud Z, Springmeyer S, et al. Clinical utility of a plasma protein classifier for indeterminate lung nodules. Lung 2015;193:1023-7. [Crossref] [PubMed]
- Fahrmann JF, Grapov D, DeFelice BC, et al. Serum phosphatidylethanolamine levels distinguish benign from malignant solitary pulmonary nodules and represent a potential diagnostic biomarker for lung cancer. Cancer Biomark 2016;16:609-17. [Crossref] [PubMed]
- Silvestri GA, Tanner NT, Kearney P, et al. Assessment of plasma proteomics biomarker's ability to distinguish benign from malignant lung nodules: results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) trial. Chest 2018;154:491-500. [Crossref] [PubMed]
- Trivedi NN, Arjomandi M, Brown JK, et al. Risk assessment for indeterminate pulmonary nodules using a novel, plasma-protein based biomarker assay. Biomed Res Clin Prac 2018. doi: 10.15761/BRCP.1000173. [Crossref]
- Ajona D, Okrój M, Pajares MJ, et al. Complement C4d-specific antibodies for the diagnosis of lung cancer. Oncotarget 2017;9:6346-55. [Crossref] [PubMed]
- Du Q, Yu R, Wang H, et al. Significance of tumor-associated autoantibodies in the early diagnosis of lung cancer. Clin Respir J 2018;12:2020-8. [Crossref] [PubMed]
- Yang D, Zhang X, Powell CA, et al. Probability of cancer in high-risk patients predicted by the protein-based lung cancer biomarker panel in China: LCBP study. Cancer 2018;124:262-70. [Crossref] [PubMed]
- Kammer MN, Kussrow AK, Webster RL, et al. Compensated interferometry measures of CYFRA 21-1 improve diagnosis of lung cancer. ACS Comb Sci 2019;21:465-72. [Crossref] [PubMed]
- Lastwika KJ, Kargl J, Zhang Y, et al. Tumor-derived autoantibodies identify malignant pulmonary nodules. Am J Respir Crit Care Med 2019;199:1257-66. [Crossref] [PubMed]
- Shen J, Liu Z, Todd NW, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer 2011;11:374. [Crossref] [PubMed]
- Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev 2013;22:540-8. [Crossref] [PubMed]
- Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. [Crossref] [PubMed]
- Gumireddy K, Li A, Chang DH, et al. AKAP4 is a circulating biomarker for non-small cell lung cancer. Oncotarget 2015;6:17637-47. [Crossref] [PubMed]
- Montani F, Marzi MJ, Dezi F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063. [Crossref] [PubMed]
- Barón AE, Kako S, Feser WJ, et al. Clinical utility of chromosomal aneusomy in individuals at high risk of lung cancer. J Thorac Oncol 2017;12:1512-23. [Crossref] [PubMed]
- Xi K, Wang W, Wen Y, et al. Combining plasma miRNAs and computed tomography features to differentiate the nature of pulmonary nodules. Front Oncol 2019;9:975. [Crossref] [PubMed]
- Molina R, Filella X, Augé JM, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol 2003;24:209-18. [Crossref] [PubMed]
- Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012;76:138-43. [Crossref] [PubMed]
- Bornhop DJ, Kammer MN, Kussrow A, et al. Origin and prediction of free-solution interaction studies performed label-free. Proc Natl Acad Sci U S A 2016;113:E1595-604. [Crossref] [PubMed]
- Kammer M, Kussrow A, Carter MD, et al. Rapid quantification of two chemical nerve agent metabolites in serum. Biosens Bioelectron 2019;131:119-27. [Crossref] [PubMed]
- Kammer MN, Kussrow A, Gandhi I, et al. Quantification of opioids in urine using an aptamer-based free-solution assay. Anal Chem 2019;91:10582-8. [Crossref] [PubMed]
- Kearney P, Hunsucker SW, Li XJ, et al. An integrated risk predictor for pulmonary nodules. PLoS One 2017;12:e0177635. [Crossref] [PubMed]
- Al Nasrallah N, Sears CR. Biomarkers in pulmonary nodule diagnosis: is it time to put away the biopsy needle? Chest 2018;154:467-8. [Crossref] [PubMed]
- Massion PP, Healey GF, Peek LJ, et al. Autoantibody signature enhances the positive predictive power of computed tomography and nodule-based risk models for detection of lung cancer. J Thorac Oncol 2017;12:578-84. [Crossref] [PubMed]
- Edelsberg J, Weycker D, Atwood M, et al. Cost-effectiveness of an autoantibody test (EarlyCDT-Lung) as an aid to early diagnosis of lung cancer in patients with incidentally detected pulmonary nodules. PLoS One 2018;13:e0197826. [Crossref] [PubMed]
- Lee JR, Chan CT, Ruderman D, et al. Longitudinal monitoring of antibody responses against tumor cells using magneto-nanosensors with a nanoliter of blood. Nano Lett 2017;17:6644-52. [Crossref] [PubMed]
- Fish A, Vachani A, Massion P, et al. Novel Multiplexed plasma biomarkers and clinical factors augment risk assessment for indeterminate pulmonary nodules in former smokers. Am J Respir Crit Care Med 2019;199:A7452.
- Jeong JS, Jiang L, Albino E, et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteomics 2012;11:O111.016253.
- Rho JH, Lampe PD. High-throughput analysis of plasma hybrid markers for early detection of cancers. Proteomes 2014;2:1-17. [Crossref] [PubMed]
- Deppen SA, Massion PP, Blume J, et al. Accuracy of a novel histoplasmosis enzyme immunoassay to evaluate suspicious lung nodules. Cancer Epidemiol Biomarkers Prev 2019;28:321-6. [Crossref] [PubMed]
- Maiga AW, Deppen S, Scaffidi BK, et al. Mapping histoplasma capsulatum exposure, United States. Emerg Infect Dis 2018;24:1835-9. [Crossref] [PubMed]
- Showe MK, Vachani A, Kossenkov AV, et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res 2009;69:9202-10. [Crossref] [PubMed]
- Wrangle J, Machida EO, Danilova L, et al. Functional identification of cancer-specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer. Clin Cancer Res 2014;20:1856-64. [Crossref] [PubMed]
- Silvestri GA, Vachani A, Whitney D, et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med 2015;373:243-51. [Crossref] [PubMed]
- Ding C, Zhou X, Xu C, et al. Circulating tumor cell levels and carcinoembryonic antigen: An improved diagnostic method for lung adenocarcinoma. Thorac Cancer 2018;9:1413-20. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Aldrich MC, Mercaldo SF, Sandler KL, et al. Evaluation of USPSTF lung cancer screening guidelines among African American adult smokers. JAMA Oncol 2019;5:1318-24. [Crossref] [PubMed]
- Guida F, Sun N, Bantis LE, et al. Assessment of lung cancer risk on the basis of a biomarker panel of circulating proteins. JAMA Oncol 2018;4:e182078. [Crossref] [PubMed]
- Vlahos I, Stefanidis K, Sheard S, et al. Lung cancer screening: nodule identification and characterization. Transl Lung Cancer Res 2018;7:288-303. [Crossref] [PubMed]
- Manning D, Ethell S, Donovan T, et al. How do radiologists do it? The influence of experience and training on searching for chest nodules. Radiography 2006;12:134-42. [Crossref]
- van Riel SJ, Jacobs C, Scholten ET, et al. Observer variability for Lung-RADS categorisation of lung cancer screening CTs: impact on patient management. Eur Radiol 2019;29:924-31. [Crossref] [PubMed]
- Singh S, Pinsky P, Fineberg NS, et al. Evaluation of reader variability in the interpretation of follow-up CT scans at lung cancer screening. Radiology 2011;259:263-70. [Crossref] [PubMed]
- Balagurunathan Y, Schabath MB, Wang H, et al. Quantitative imaging features improve discrimination of malignancy in pulmonary nodules. Sci Rep 2019;9:8528. [Crossref] [PubMed]
- Nasrullah N, Sang J, Alam MS, et al. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors (Basel) 2019. [Crossref] [PubMed]
- Gupta A, Saar T, Martens O, et al. Automatic detection of multisize pulmonary nodules in CT images: large-scale validation of the false-positive reduction step. Med Phys 2018;45:1135-49. [Crossref] [PubMed]
- Nishio M, Nagashima C. Computer-aided diagnosis for lung cancer: usefulness of nodule heterogeneity. Acad Radiol 2017;24:328-36. [Crossref] [PubMed]
- Armato SG 3rd, Drukker K, Li F, et al. LUNGx challenge for computerized lung nodule classification. J Med Imaging (Bellingham) 2016;3:044506. [Crossref] [PubMed]
- Cherezov D, Hawkins SH, Goldgof DB, et al. Delta radiomic features improve prediction for lung cancer incidence: a nested case-control analysis of the National Lung Screening Trial. Cancer Med 2018;7:6340-56. [Crossref] [PubMed]
- Kadir T, Arteta C, Pickup L, et al. Solid and part-solid lung nodule classification using deep learning on the national lung screening trial dataset. Am J Respir Crit Care Med 2018;197:A7417.
- Kadir T, Arteta C, Pickup L, et al. Deep learning based risk stratification of patients with suspicious nodules. Am J Respir Crit Care Med 2018;197:A4695.
- Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. [Crossref] [PubMed]
- Massion PP, Antic S, Ather S, et al. Assessing the Accuracy of a Deep Learning Method to Risk Stratify Indeterminate Pulmonary Nodules. Am J Respir Crit Care Med 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Huang P, Lin CT, Li Y, et al. Prediction of lung cancer risk at follow-up screening with low-dose CT: a training and validation study of a deep learning method. The Lancet Digital Health 2019;1:e353-62. [Crossref]
- Gould MK, Donington J, Lynch WR, et al. Evaluation of individuals with pulmonary nodules: when is it lung cancer? Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e93S-120S.
- Kerr KF, Marsh TL, Janes H. The importance of uncertainty and opt-in v. opt-out: best practices for decision curve analysis. Med Decis Making 2019;39:491-2. [Crossref] [PubMed]
- Kerr KF, Brown MD, Marsh TL, et al. Assessing the clinical impact of risk models for opting out of treatment. Med Decis Making 2019;39:86-90. [Crossref] [PubMed]
- Ost DE, Gould MK. Decision making in patients with pulmonary nodules. Am J Respir Crit Care Med 2012;185:363-72. [Crossref] [PubMed]
- Deppen SA, Grogan EL. Using clinical risk models for lung nodule classification. Semin Thorac Cardiovasc Surg 2015;27:30-5. [Crossref] [PubMed]
- Pepe MS, Kerr KF, Longton G, et al. Testing for improvement in prediction model performance. Stat Med 2013;32:1467-82. [Crossref] [PubMed]
- Paynter NP, Cook NR. A bias-corrected net reclassification improvement for clinical subgroups. Med Decis Making 2013;33:154-62. [Crossref] [PubMed]
- Paynter NP, Cook NR. Adding tests to risk based guidelines: evaluating improvements in prediction for an intermediate risk group. BMJ 2016;354:i4450. [Crossref] [PubMed]
- Cook NR, Ridker PM. Advances in measuring the effect of individual predictors of cardiovascular risk: the role of reclassification measures. Ann Intern Med 2009;150:795-802. [Crossref] [PubMed]
- Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 2007;115:928-35. [Crossref] [PubMed]


