
Novel insights into the molecular basis of calcific aortic valve disease
Calcific aortic valve disease (CAVD) is the most common valvular heart disease in the elderly. CAVD is a slowly progressive disorder that ranges from mild valve thickening without obstruction of blood flow, termed aortic sclerosis, to severe calcification with impaired leaflet motion, termed aortic stenosis (AS). CAVD is in one of the most important contributors to cardiovascular morbidity and mortality (1,2). AS occurs in 2% of people above 65 years old, and its prevalence is likely to increase due to the aging of the population (3). Although many studies have been made to understand the factors driving progression of CAVD, the molecular mechanisms and signalling pathways underlying the pathophysiological responses of this disease are still obscure.
Pathogenesis of CAVD
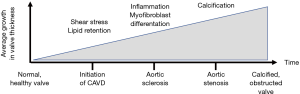
CAVD is recognized as an actively regulated disease (4). In the initiation phase, risk factors it shares with atherosclerosis and mechanical strain contribute to the endothelial damage on the aortic valve, leading to infiltration of oxidized phospholipids (OxPL) into the valve (Figure 1). Both the endothelial dysfunction and the retention of OxPL promote inflammatory infiltrates to accumulate into the valve (5) that in turn promote profibrotic processes, stimulating myofibroblastic differentiation of valve interstitial cells (VICs) and extracellular matrix (ECM) modelling. Eventually, cellular pathways that promote calcification are activated, and calcification starts to accumulate leading to progressive aortic valve obstruction (6).
Relevance of omics studies in CAVD
Generation of “omics data” via high-throughput technologies such as DNA microarrays and proteomics have provided a deeper understanding of biological processes, molecular pathways, and interaction networks involved in cardiovascular diseases (7). As the pathobiology of CAVD is the sum of several contributing processes, omics-analyses have been used to understand the complexity of the disease progression. Several studies have compared transcriptomic (8-10) and proteomic (11-13) profiles of human calcified and normal aortic valves. However, the studies have often a small sample size, and also different technology platforms and data processing techniques have been used in producing altogether inconsistent results. In their study, Zhang and Ma (in 2020) used an integrated bioinformatics analysis to overcome this problem of heterogenicity (14). They combined gene expression profiling datasets, performed functional and pathway enrichment analysis and constructed the protein-protein interaction (PP1) networks to reveal key genes and molecular mechanisms that contribute to the progression of CAVD.
Key genes and pathways in CAVD
The study of Zhang and Ma (in 2020) identified altogether 53 up- and 54 down-regulated genes in CAVD by integrated analysis of two datasets (8,10). Since individual genes are often involved in multiple cellular processes, the authors identify also the molecular processes associated to dysregulated genes highlighting the importance on ECM modelling and immune response related processes in CAVD.
They further identified hub of six genes in protein-protein interaction (PPI) network analysis. These genes, VCAM1, MMP9, ITGB2, RAC2, vWF and ALDH2, were mainly enriched in cell adhesion and leukocyte transendothelial migration pathways. The potential significance of key genes for disease progression of CAVD is discussed by the authors, but it is notable that so many of them is contributing to inflammatory response. Confirming validity of their study, the roles of vascular cell adhesion molecule 1 (VCAM1) and matrix metalloproteinase 9 (MMP9) are well defined in CAVD (15,16). VCAM1 is expressed in the endothelial cells and facilitates inflammatory cell infiltration into the valvular tissue whereas MMP9 along with the other MMPs and tissue inhibitors of metalloproteinases (TIMPs) promote ECM remodelling even in an early phase of CAVD and the fibrotic process continues also in the advanced stages of the disease (6,17). von Willebrand factor (VWF) is a glycoprotein that interacts with several factors that are associated with AS including ADAMTS13, Factor VIII, collagens and integrins (18). VWF has an important role in platelet adhesion and hemostasis and it might contribute to the regulation of angiogenesis (18). The intriguing finding of the study of Zhang and Ma (2020) is that among the key genes are integrin beta 2 (ITGB2), Rac family small GTPase 2 (Rac2) and aldehyde dehydrogenase (ALDH2) but no previous research has investigated their role in CAVD. Integrins have been shown to enhance migration of leukocytes into the tissues and they could act also as mechanosensor on the cell surface of VICs in response to increased shear stress (19). The future studies will show whether Rac2 and ALDH2 play a role in CAVD.
Lastly, the authors generated a PPI network that is based on previously known interactions among the identified genes. It is important to note that this is a putative network, and these interactions have not been confirmed to happen in aortic valves. In fact, based on transcriptomics we might have a complete picture on what RNAs are produced within aortic valve, but it is not known if these genes are translated to functional proteins. However, since the impact of individual gene on the disease phenotype is often modest in complex diseased like CAVD, these interaction networks may shed some light on the complicated disease process. Concequently, the next step will be to confirm the expression of these genes with other quantitative gene expression platform and to perform additional experiments elucidate the interactions between dysregulated genes.
In conclusion, the progression from normal valve to aortic sclerosis and further to AS is slow. Progressive stenosis occurs in almost all patients, even though there is large variation in the speed of disease progression in individual patients. The present study expands the overall data on CAVD transcriptomics by performing integrated data analysis of gene expression changes in order to better understand molecular drivers contributing to the progression of CAVD. Consequently, this data may be useful in developing therapeutic strategies to treat this disease.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Thoracic Disease. The article did not undergo external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1669). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lindman BR, Clavel MA, Mathieu P, et al. Calcific aortic stenosis. Nat Rev Dis Prim 2016;2:16006. [Crossref] [PubMed]
- Otto CM, Prendergast B. Aortic-valve stenosis - From patients at risk to severe valve obstruction. N Engl J Med 2014;371:744-56. [Crossref] [PubMed]
- Stewart BF, Siscovick D, Lind BK, et al. Clinical factors associated with calcific aortic valve disease. J Am Coll Cardiol 1997;29:630-4. [Crossref] [PubMed]
- Rajamannan NM, Evans FJ, Aikawa E, et al. Calcific aortic valve disease: Not simply a degenerative process: A review and agenda for research from the national heart and lung and blood institute aortic stenosis working group. Circulation 2011;124:1783-91. [Crossref] [PubMed]
- Bochkov VN, Oskolkova OV, Birukov KG, et al. Generation and biological activities of oxidized phospholipids. Antioxid Redox Signal 2010;12:1009-59. [Crossref] [PubMed]
- Dweck MR, Boon NA, Newby DE. Calcific aortic stenosis: A disease of the valve and the myocardium. J Am Coll Cardiol 2012;60:1854-63. [Crossref] [PubMed]
- Leon-Mimila P, Wang J, Huertas-Vazquez A. Relevance of Multi-Omics Studies in Cardiovascular Diseases. Front Cardiovasc Med 2019. [Crossref] [PubMed]
- Guauque-Olarte S, Droit A, Tremblay-Marchand J, et al. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol Genomics 2016;48:749-61. [Crossref] [PubMed]
- Bossé Y, Miqdad A, Fournier D, et al. Refining molecular pathways leading to calcific aortic valve stenosis by studying gene expression profile of normal and calcified stenotic human aortic valves. Circ Cardiovasc Genet 2009;2:489-98. [Crossref] [PubMed]
- Ohukainen P, Syvaranta S, Napankangas J, et al. MicroRNA-125b and chemokine CCL4 expression are associated with calcific aortic valve disease. Ann Med 2015;47:423-9. [Crossref] [PubMed]
- Martín-Rojas T, Gil-Dones F, Lopez-Almodovar LF, et al. Proteomic profile of human aortic stenosis: Insights into the degenerative process. J Proteome Res 2012;11:1537-50. [Crossref] [PubMed]
- Martin-Rojas T, Mourino-Alvarez L, Alonso-Orgaz S, et al. ITRAQ proteomic analysis of extracellular matrix remodeling in aortic valve disease. Sci Rep 2015;5:17290. [Crossref] [PubMed]
- Schlotter F, Halu A, Goto S, et al. Spatiotemporal multi-omics mapping generates a molecular atlas of the aortic valve and reveals networks driving disease. Circulation 2018;138:377-93. [Crossref] [PubMed]
- Zhang Y, Ma L. Identification of key genes and pathways in calcific aortic valve disease by bioinformatics analysis. J Thorac Dis 2019;11:5417-26. [Crossref] [PubMed]
- Mazzone A, Epistolato MC, De Caterina R, et al. Neoangiogenesis, T-lymphocyte infiltration, and heat shock protein-60 are biological hallmarks of an immunomediated inflammatory process in end-stage calcified aortic valve stenosis. J Am Coll Cardiol 2004;43:1670-6. [Crossref] [PubMed]
- Perrotta I, Sciangula A, Aquila S, et al. Matrix metalloproteinase-9 expression in calcified human aortic valves: A histopathologic, immunohistochemical, and ultrastructural study. Appl Immunohistochem Mol Morphol 2016;24:128-37. [Crossref] [PubMed]
- Fondard O, Detaint D, Iung B, et al. Extracellular matrix remodelling in human aortic valve disease: The role of matrix metalloproteinases and their tissue inhibitors. Eur Heart J 2005;26:1333-41. [Crossref] [PubMed]
- Van Belle E, Vincent F, Rauch A, et al. von Willebrand Factor and Management of Heart Valve Disease: JACC Review Topic of the Week. J Am Coll Cardiol 2019;73:1078-88. [Crossref] [PubMed]
- Rutkovskiy A, Malashicheva A, Sullivan G, et al. Valve interstitial cells: The key to understanding the pathophysiology of heart valve calcification. J Am Heart Assoc 2017;6:1-23. [Crossref] [PubMed]


