Study on automatic detection and classification of breast nodule using deep convolutional neural network system
Introduction
Breast cancer is one of the most common malignancies worldwide and a leading cause of cancer death in female. Worldwide, as it was analyzed in Global cancer statistics of 2012, breast cancer affected more people than any other cancer, accounting for approximately 23% of the total number of cancer cases and 14% of cancer deaths (1).
Ultrasound (US) is used as an attractive screening tool and first-line diagnostic tool for breast cancer because of its well recognized advantages of real-time images, prompt reporting, relatively low cost, readily availability and, in particular, non-invasion, no radiation (2-4). Even more importantly, US showed high sensitivity in detecting dense breast nodule, which is believed to be commonly existed in Asian female and more likely to develop into breast cancer (5). Images of Automated Breast Ultrasound (ABUS) are acquired by a motor-driven transducer, which scan breasts automatically. ABUS has several attractive advantages over conventional hand-held US, such as higher reproducibility, less operator-dependence, fewer physician time for image acquisition (6), standardized US documentation, and abundant diagnostic information of malignancy obtained from the reconstructed coronal planes (7). Nevertheless, there are several factors that may make breast US diagnosis difficult, especially when apply ABUS (8). Firstly, the poor image resolution between nodule and the surrounding breast tissue may make the detection task ineffective, especially when the nodule exhibit a vague margin in images. Secondly, the nodules are relatively small in size compared with surrounding background breast tissue, which would prevent the reader from focusing on useful information. Arleo et al. (9) found the recall rate (also known as sensitivity) of screening with ABUS ranged from 24.7% to 12.6%, suggesting readers’ experience, in other words, human error, should much responsible for the inaccuracy of ABUS diagnosis. What’s worse, as a large amount of images are produced by ABUS, manually image reading by physician is time-consuming. To overcome these limitations, a concept of computer-aided diagnosis (CAD) system based on the high-performance methodology of machine learning (ML) was introduced (10-13). In recent years, CAD has been experimentally used in detection and diagnosis of breast nodules. The methodology ranged from traditional ML algorithms [watershed, thresholding and clustering (5,14-16)] to recently emerged deep learning (DL) methods [convolution neural network (CNN), deconvolution neural network (12,13,17,18)]. Under the standard of less than five false positive nodule per scan or per volume, the reported sensitivity ranged from 64% to 89.19% (5,12-14,16), indicating a clinically acceptable but potentially improved sensitivity. Part of the reason for misdiagnosis is that the imaging features of breast lesions, especially in the early stage of tumorigenesis, are sometimes atypical, making it difficult to distinguish malignant from benign. Nevertheless, early and accurate diagnosis is very important for individualized therapy, because the invasion is less, the cost is lower, and the prognosis is better (19).
Compared to its predecessors, CNN may show some advantages to process atypical image in the diagnosis of breast lesions. CNN can extract the local information between image pixels with convolutional layer. Meanwhile, it has strong ability to extract the global information of the images by increasing the receptive field of the network. It has been already used for classification task in many areas of medical researches (20). Image segmentation (in our study, that is the distinguishing of nodule from surrounding normal tissues) is an essential prerequisite for the classification of benign and malignant nodule. Herein, we proposed a fast and effective three-dimensionally U-shaped convolutional neural network (3D U-Net) in detection task. To the best of our knowledge, 3D U-Net have not yet been reported for the diagnosis of breast nodules on ABUS images in published English literature. According to reported study (21) and our previous experiment of U-Net in detection of breast nodule, U-Net not only retains the advantages of CNN, but also can directly input the whole large size ABUS image for segmentation. It can obtain accurate detection results by using the context information between the images with fewer parameters, faster operation speed.
In this study, we applied ABUS to obtain the US images. Afterwards, we established a novel CAD system, which consisted a 3D U-Net for detection, and a multiple layer CNN for classification of breast nodules. The purpose of this study was to achieve the automatic US diagnosis of breast nodules, thus improve the efficiency of breast US diagnosis.
Methods
Patient and Controls Enrollment
From June 2018 to May 2019, 194 consecutive female patients and 70 negative control patients without breast nodules were retrospectively enrolled in the First Affiliated Hospital of Xi'an Jiaotong University. A total of 293 breast nodules were collected for case group.
Patients in case group who met the following inclusion criteria were enrolled in this study: (I) all these nodules of the patients were newly discovered and untreated; (II) the patients had undertaken ABUS scan before US guided biopsy of target breast nodules within one month; (III) definite pathological diagnosis of benign and malignant could be obtained by specimen from biopsies or surgical resection; (IV) the image quality of ABUS examination was good enough to show the entire margin of the lesion, no matter distinct or indistinct. The exclusion criteria of patient in case group were as follows: (I) non-nodular breast disease; (I) as the nodule was too large in size, too superficial in location or too hard in texture, the ABUS artifact was obvious and the quality of images was poor; (III) ABUS examination was not available because of skin defects and/or unbearable pain in certain breast region (such as ulcers or wounds); (IV) due to the limited amount of tissue obtained by biopsy or the atypical benign and malignant features under microscope, it is impossible to make a definite diagnosis of benign or malignant histopathologically; (V) patients received chemotherapy, radiation therapy or surgical local resection before ABUS scan, which was supposed to make the US images and histology pictures complex or atypical, thus difficult to diagnosis.
The negative controls were selected from patients with other diseases (patients hospitalized because of other disease, such as diabetes, hypertension and nephritis). ABUS showed no breast abnormalities. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent to engage in the study was obtained from each participant. The study was approved by the Ethics Committee in The First Affiliated Hospital of Xi’an Jiaotong University.
Pathologic examination
All of the 293 breast nodules underwent an US guided biopsy, which used a 16-gauge needle and a Magnum automatic biopsy gun (Bard Inc., Murray Hill, NJ, USA) with 3–6 tissue cores. All of the 176 malignant tumors underwent further extensive excision. In our study, the CAD diagnoses of 293 breast nodules were compared with the histopathological results from biopsy or surgical specimens which were taken as the gold standard.
To avoid inter-observer bias affecting the pathological diagnosis of breast nodules, the specimen were randomly assigned to three senior pathologists with over 10 years of experience in breast pathology. They independently review the histology slides and made a diagnosis.
Obtaining and pre-processing of ultrasonic images
All the data obtained in this study were completed by an ABUS Scanner (GE, model: INVENIA ABUS LQGI, USA). ABUS consists of a scanning station and a workstation. Operations in scanning station were as follows: the patient lied down on her back and raised her hands above her head. The technician smeared the coupling agent on the probe and adjusted the position of the manipulator until the coupling agent was in full contact with the probe and the skin. The technician input the patient information into the operation interface, set the position of scanning surface and the scanning times, and started the automatic scanning. The ABUS probe moved from one side to the other in the probe frame. According to the breast size, scan was performed in the medial, middle, lateral or even outermost position of the breast. The scanning criterion was to ensure that every two adjacent scanning positions overlap locally to prevent missed scans. The obtained scanned images (Figure 1A) were transmitted to the workstation in real time, where the coronal and three-dimensional images were automatically reconstructed.

The original images examined by ABUS had different resolutions and the number of pixels in each dimensions, which was 0.504 mm/pixels in transverse plane, 0.082 mm/pixels in longitudinal plane and 0.200 mm/pixels in coronal plane respectively. Therefore, we pre-processed the images before inputting to the U-Net model we established in the following steps. We adjusted the pixel spacing in the three dimensions to 0.5 mm/pixels, so that the shapes of the whole breast in the images were consistent with the actual proportion.
Manual annotation
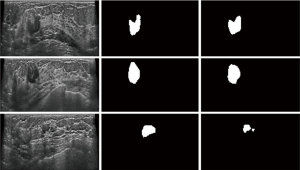
According to the proportion of 7:1:2, the pre-processed images from 293 nodules of 194 patients and 70 negative controls were randomly assigned to training set, verification set and test set. The images enrolled in the three sets were used for construction, optimization and effect verification of CNN model, respectively. The images enrolled in the training set were independently analyzed by two sonologists who have more than 5 years’ experiences in US diagnosis. Using ITK-SNAP software (http://www.itksnap.org/pmwiki/pmwiki.php?n=Main.HomePage), they manually traced the margin of nodules (the so-called region of interest), layer after layer of the images. By this way, a reference standard of 3D label was obtained (Figure 1B). This ground truth was used as a learning sample for training 3D U-Net to do segmentation tasks in the following steps.
Establishment of 3D U-Net model for detection and Deep CNN model for classification
Basic model framework
Pre-processed images from 205 breasts of case group and 49 normal breasts of negative controls were used in building the training set. After multi-layer convolution, pooling, nonlinear activation and other operations in U-Net network structure, the output of the network was the probability map which showed the category of each pixel. With the 3D U-Net model, we determined the location and margin of the nodule. Afterwards, with the CNN model we described in afterward "Model verification and evaluation" section, we computed the probability of malignancy of the nodule. The aim was to obtain the clinically significant nodule detection (Figure 1C). The results of nodular detection by detection model and the probability of malignancy by classification model were compared with the results of manual annotation by sonologist in our previously described “Obtaining and pre-processing of ultrasonic images” section, and golden standard of pathological diagnosis (Figure 1D), respectively.
Model optimization
In order to make the model more suitable for processing breast US and ABUS data, we added the following improvements to the network structure (Figure 2).
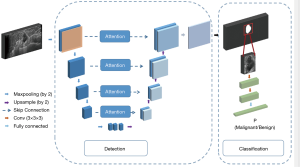
To solve the problem caused by the serious non-uniformity area between the nodules and surroundings, we introduced the spatial attention connection mechanism (22) to calculate the low-level information in the encoder and obtain a new feature map. It was connected in series with the feature map on the right side of the U-Net model, to utilize the fine-grain features properly.
To reduce the risk of the “false nodules” detected in the blinding area for scanning, such as edge of the muscle layer, fat layer and nearby armpit, we analyzed the characteristics of the global anatomical structure carefully. Specifically, we applied CoordConv (23) in the first layer of this detection model, in which case, we calculated relative coordinates of each pixel on the images and concatenated them with the input image. In this way, we can add the spatial position information to this model.
To solve the problem of gradient vanishing, we replaced the simple stacked convolution layer in basic U-Net with residual block. Very deep network can represent complex functions. However, a huge barrier to training them is vanishing gradients. Very deep networks usually have a gradient signal that goes to zero quickly, thus making the decline of gradient prohibitively slow. In this regard, we added the shortcut connections and bottleneck architecture. By learning a function that is easier to identify, we aimed to help the model address the problem of vanishing gradient.
Due to the severe data imbalance between nodule region and non-nodule regions, some non-nodular pixels in the image would be mistakenly marked, while the nodular pixels would be missed. Through the hard mining of all the positive and negative cases, the mismarked pixels would be detected and thus be excluded. They would not have the chance to be used for the calculation of the loss function in the following steps. Hard mining would prevent the model from learning the wrong information.
To solve the problem that it is difficult for a single research center (our hospital) to access to a large amount of data in a limited research period, some simple and practical processes were adopted for data augmentation. We have carried out operations of flipping horizontally and vertically, position-translation transformation, scaling, shearing, rotation and contrast adjustment, effectively increasing the amount of training data.
Model verification and evaluation
On the test set, in order to compare the effective of the models in this study with those reported in the literature, we put the verification set of ABUS imaging data in this study (59 lesions and 14 negative controls, according to 7:1:2 ratio) into some previous researched model(watershed, FCN, V-Net, PSPNet and basic 3D U-Net), to get the diagnostic efficiency (sensitivity, specificity, accuracy). By doing this, the efficiency of our preliminary detection model was compared with those of the previous methods. The images of 29 case and seven controls were input into the model, which was established in the “Manual annotation” section, in order to compare the sensitivity and specificity of preliminary model and the previous ones in literature. Moreover, in the process of model training, we screen out the difficult samples according to the error between the label of the training sample and the network prediction value. The greater the error, the more difficult it is to detect the sample. In this way, the difficult samples are screened out, and then in the training process, we focus on sending these difficult samples into the network for many times of training. At the end of this step, a final model with maximum optimization had been determined.
On the verification set, the detection efficiency of our model was evaluated. Specifically, we input the images of 59 nodules and 14 negative breasts into final model. Both true and false negative rate of nodule detected in each image were statistically analyzed. Intersection over Union (IOU) of our study was to evaluate the coincidence between the predicted and actual target nodules of all 293 cases. Sensitivity and IOU of the detection model were calculated in python.
Classification model
We input the images of cases which IOU were more than 70% in all three directions (267 nodules in total) to a CNN. The CNN consisted of two convolutional layers with batch normalization and ReLU activation. At the end of the network, a softmax layer was used after the fully connected layer to map the outputs into classification probabilities. Taking the benign and malignant results of histopathological diagnosis as the reference standard, true positive rate and false positive rate of malignancy of the classification model were calculated. Sensitivity, specificity and accuracy of the classification model were calculated in python. Receiver operating characteristic (ROC) curve of our classification network was plot accordingly in Python.
Formula: IOU =A∩B/A∪B, (In our study, A and B refer to the area of actual nodule and “predicted” nodule by the segmentation model.) Sensitivity (Recall) = True Positive / (False Negative + True Positive) Specificity = True Negative / (True Negative + False Positive) Accuracy = (True Positive + True Negative) / (Positive + Negative)
Results
The baseline characteristics of the study population were as follows. Mean diameter of 293 nodules was 1.87±0.83 cm. Median age of 293 patients was 54.31±9.68 years (range, 37–75 years). Malignancy (176 cases) included invasive ductal carcinoma (97cases), invasive lobular carcinoma (35 cases), invasive papillary carcinoma (25 cases), ductal carcinoma in situ (14 cases), mucinous carcinoma (three cases), and unclassified type cancer (two cases). Benign (117 cases) included fibroadenoma (67 cases), inflammation (15 cases), sclerosing adenosis (9 cases), fibroadenomatoid hyperplasia (9 cases), fibrocystic change (7 cases), papilloma (5 cases), and pseudoangiomatous stromal hyperplasia (5 cases).
One hundred artificially marked nodules were randomly selected, and the tumor areas of breast nodules segmented by two sonologists were compared by two independent samples t test. It was found that there was no significant difference in manual segmentation area between the two doctors (P>0.05). The results revealed that the heterogeneity of manual segmentation performance by the two doctors was small. Therefore, the manual annotation could be used as a reliable reference for subsequent training automatic segmentation model.
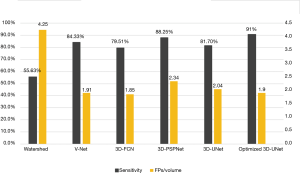
Our optimized 3D U-Net had a detection sensitivity of 91% and false negative number of 1.92 FPS per ABUS volume. The sensitivity was highest while false negative number was at a lower level when compared with previous methods (Figure 3).
IOU of our segmentation model was 70% in three directions (transverse, longitudinal, and coronal views). For our proposed CNN model, 91% of the nodules yielded a total IOU of more than 70% in the whole volume (Figure 4).
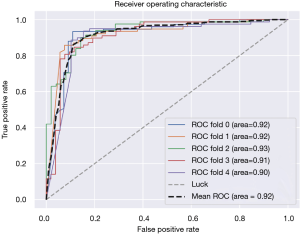
The followed CNN classifier also showed promising performance for classification of breast tumors, with an acceptable sensitivity, specificity and accuracy of 87%, 88% and 87.5%, respectively. The area under the ROC curve (AUC) was 0.922 with the 95% confidence interval between 0.893 and 0.935 (Figure 5).
Discussion
As main complementary examination to mammography, US was much superior to mammography due to its attractive advantages including, but not limited to, high sensitivity especially in dense breasts, are unexposed to X-rays and are safe for young, pregnant, lactating population (28,29). Nevertheless, conventional US manual scanning of breast have some limitations such as operator dependency and poor reproducibility. What is worse, simultaneous gathering and interpretation of images made radiologists exhausted and may increase the missing rate of cancers (28). ML is a sub-field of artificial intelligence, which allows computers to learn from data without being explicitly programmed (30), and shows promising advantages in fast, accurate, friendly use and low-cost detection of targets in medical imaging. However, the applications of artificial intelligence and ML in US field were far less than in magnetic resonance and computed tomography field (30). This maybe due to the fact that the images produced by real-time performance of US, different doctors and different US machine units, are difficult to be standardize and stored. Nevertheless, excitingly, ABUS machine, which can scan automatically, standardize and transfer numerous amounts of data to computers, solved this problem to a great extent. More importantly, ABUS can provide more diagnostic information regarding malignancy by the reconstructed coronal planes. The most remarkable feature obtained by ABUS is retraction phenomenon, which has been proved to be a crucial feature of breast malignant tumor with high specificity (7). It is automatically reconstructed by an ABUS machine based on the automatically scanned sagittal and cross-section images. That’s why in this study, we used ABUS to obtain data and the technology of ML to analyze data.
In recent years, some studies have applied ML to the detection and diagnosis of breast nodules. In particular, some commercial software developed specifically for breast CAD (11) demonstrated the successful clinical application of ML in breast CAD. However, the accuracy in related studies needed to be improved. This is partly because steps of traditional ML methods are cumbersome or inaccurate to some extent. The methodology of ML are divided into DL methods, which has emerged since the 2010s and traditional ML techniques. For classification tasks, most traditional methods have limited ability to process natural data in the original form. It required careful engineering and considerable domain expertise to design a feature extractor (20). Noise reduction, filtering and other complex pre-processing and are required as well. A number of complex algorithms in the field of computer vision have been proposed for this purpose and it is necessary to extract varies information of nodule locations, sizes, shapes, densities, and other features (31). This process is largely arbitrary and time-consuming because it may run the risk of including useless and redundant features and, more importantly, may not include truly useful features (32). All these disadvantages of traditional methods heavily limited the application in practical medical work. An image is made up of millions of pixels. Pixel-level features make no sense for the features of the entire image. In that case, through complex and structured processing of pixels, CNN directly automatically extract the deep-seated features of the entire image without any human supervision. CNN, a recently emerged but popular algorithm of DL method, have been applied with great success to the detection, segmentation and recognition of objects and regions in images and now become the dominant approach for almost all recognition and detection tasks (20). The Deep CNNs were inspired by the human visual system, which enabled certain properties to be encoded and reduced the strong dependence of domain knowledge (33). It does not require a complex pre-processing process, nor human engineers to use the complex professional background knowledge to manually design and select features (20). Compared with the traditional methods, it has shorter operation time and better generalization (34). Impressively, it has beaten traditional ML techniques in the strong learning and processing ability of large even huge amounts of data. CNN was considered to be suitable for processing breast data, especially those generated by ABUS (35). The convolution layer, nonlinear layer and pooled layer of CNN can learn the global information of image pixels, and thus to extract the high-level features hidden inside the images. U-Net is a modified and extended version of a fully convolutional network, which has been widely used in the tasks of medical image segmentation. U-Net can not only retain the above advantages of CNN, but also is especially suitable for process ABUS images. This is due to the fact that it can directly input the large-scale ABUS images for detection task (36) without complex preprocessing. With fewer parameters and faster operation speed, U-Net can produce accurate segmentation results. Therefore, U-Net is suitable for CAD, especially when using ABUS images.
We found that compared with other organs or tissues, breast US images have some inherent characteristics, which may bring some difficulties to the application of CAD in breast, especially in the task of accurate segmentation of nodule. Firstly, the conventional 2D convolutional layers can only deal with 2D input images (37). Alarmingly, in terms of the 3D volumes obtained by ABUS scanner, single 2D slices would lose the relative context of the nodule. What is worse, the surrounding tissue is unavailable to be take into account in the segmentation of the nodule. As a result, “false nodule” can be easily detected in the non-breast region of fat layer and muscle layer. Notably, some researchers have set out to reduce the false positive rate by segmenting the breast, the nipple, and the chest-wall in 3D ABUS images. Regrettably, the reported low sensitivity (sensitivity of 64% at one false positives per image) need to be further improved (12). Secondly, the image contrast between the focus and the surrounding tissue is low while the gray scale of the whole US image is not uniform. What is worse, the nodule is generally small relative to the entire breast US image. In our study, these problems have been well solved by 3D U-Net as follows. By calculating the probability of whether each pixels in the image is a nodule, the U-Net network automatically output the segmentation prediction probability map of the whole nodule. We adopted the optimization strategy of 3D U-Net, and adding spatial position information with 3D CoordConv in the modeling. 3D U-Net can directly use the 3D volume image and their corresponding label as the training sample of the network. By taking the 3D cubes from three orthogonal planes rather than 2D image blocks as the network input, the spatial relationship of pixels can be fully utilized (38). It can obtain the accurate segmentation results by using the context information between neighbor slices (39).
There are two important aspects of CAD research associating with each other. In the “Detection” phase, the nodule is segmented from the normal tissues, while in the subsequent “Diagnosis” phase, the nodule is evaluated to produce a benign/malignant diagnosis (40). Segmenting nodules in medical images demands a higher level of accuracy than in natural images. As subtle patterns around a nodule may indicate nodule malignancy, mistakenly excluding them from the segmentation mask will reduce the credibility of the model (41), thus lead to the wrong result in “Diagnosis” phase. Introduction of the unnecessary information from the surrounding non-nodular tissue will also have a negative effect in “Diagnosis” phase. Therefore, in addition to evaluating the detection rate, the similarity of the area between “predicted” and actual nodules in the “detection” phase should also be taken into account. However, many previous CAD studies of breast US had not mentioned it in their methods while in other study the minimum value (exact data not mentioned in the article) was set as the overlapped result to consider as an effective test results (5). A few studies of breast US image segmentation used the Dice coefficient (42,43) or IOU (also known as Jaccard coefficient) (38) to evaluate segment efficiency of the breast nodules. Dice coefficient is calculated as twice the overlap of the “predicted” and actual area divided by the sums of the “predicted” and actual area. IOU is calculated as the overlap of the “predicted” and actual area divided by the union of the “predicted” and actual area. Our study used the IOU. To the best of our knowledge, few studies have evaluated the IOU of breast US segmentation by considering all three-dimensions. We defined a nodule as “detected” only if the IOU was more than 70% in all three directions (transverse, longitudinal, and coronal views). With this rigid cutoff value, we were delighted to find that, 91% of the real nodules were “detected” by our optimized model (overlap more than 70% on the image) (Figure 4). The high similarity between prediction and actual nodules provided a good guarantee for the follow-up “diagnosis” phase.
Accuracy should be a critical factor in determining the quality of tools for the diagnosis of breast nodules. Some previous studies had high positive rates, but at the expense of high false positive rates (44,45). As for benign nodule, over-diagnosis may increase anxiety of patients and their families about breast cancer, as well as the economic costs to both patients and the health care system due to additional and potentially unnecessary diagnostic tests (19). Considering high false positive rate, doctors have to carefully examine the results obtained by ML. Frankly, the existence of too many false positive results can not improve the effectiveness of doctors in reading the image data, nor can it save the doctors’ time and energy. To make matters worse, it will wipe out doctors' enthusiasm for using ML (13). Considering these issues, the importance of true negative rate and false negative rate should be fully calculated when constructing the detection model. Our optimized U-Net model was able to achieve the highest sensitivity (92.2%) than the previous models we compared in this study. Meanwhile, the number of false positives obtained by our model was lower (1.9 per volume scan) than models in previously reported works (44).
In general, the diagnosis of benign and malignant nodules depends to a large extent on the visible US features such as the boundary, edge, shape, internal echo and posterior shadow of the nodules, the results of which can be explained clinically. However, the results of the CNN model for differentiating the benign and malignant nodules are based on the pixel-wise analysis of images, which may not be clinically interpretable. This fact suggests that CNN may be able to find some high-level features in US images that can not be explained or invisible to the naked eye, which is the hot topic in radiomics research (46).
The limitations of this study were as follows. First, the dataset was relatively small. The good performance of DL depends to a large extent on the mass of big data, while small sample size may lead to over-fitting and affect the generalization of the model. Secondly, compared with proportion in clinical work, there was some deviation between the proportion of benign and malignant nodules (117 benign and 176 malignancy, respectively) in our study. This was because, in order to obtain definite histopathological results, we inevitably excluded some patients who choose follow-up plan rather than biopsy when a benign diagnosis was made by US examination of breast. Last but not the least, as our exclusion criteria described below, a few lesions were excluded because of non-nodular breast disease (17 cases) and undetectable or difficult-to-detect in ABUS (23 lesions), respectively. In other words, we cannot evaluate the diagnostic efficiency of these cases, though they account for only a small proportion of the whole patient population (compared with 293 lesions enrolled in this study).
Conclusions
This study proposes a novel model for the detection and diagnosis of breast cancer using ABUS scans. This study found that the combined application of ABUS to obtain breast US imaging data, a detection model established by an optimized 3D U-Net and a diagnostic model constructed by multiple layer CNN method would produce a high detection rate of breast nodules and an acceptable diagnostic accuracy of breast malignant tumors. To sum up, on the basis of this study, we recommend deep CNN as an alternative auxiliary means for the detection and diagnosis of breast nodules. However, if deep CNN is expected to be used in diagnosis-related clinical decision-making of breast cancer, large sample, multicenter, prospective studies should be mandatory.
Acknowledgments
Funding: This work was supported by Natural Science Foundation of Shaanxi Province (No.2018SF-245) of China, Fundamental Research Funds for the Central Universities (grant number Xjj2017qyzh07). and in part by National Key Research and Development Program (grant number. 2016YFB1000303) of China.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-19-3013). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent to engage in the study was obtained from each participant. The study was approved by the local Ethical committee of the First Affiliated Hospital of Xi’an Jiaotong University (Shanxi, China).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gu S, Xue J, Xi Y, et al. Evaluating the effect of Avastin on breast cancer angiogenesis using synchrotron radiation. Quant Imaging Med Surg 2019;9:418-26. [Crossref] [PubMed]
- Fiorica JV. Breast cancer screening, mammography, and other modalities. Clin Obstet Gynecol 2016;59:688-709. [Crossref] [PubMed]
- Yan F, Song Z, Du M, et al. Ultrasound molecular imaging for differentiation of benign and malignant tumors in patients. Quant Imaging Med Surg 2018;8:1078-83. [Crossref] [PubMed]
- Wu GG, Zhou LQ, Xu JW, et al. Artificial intelligence in breast ultrasound. World J Radiol 2019;11:19-26. [Crossref] [PubMed]
- Lo C, Shen Y, Huang C, et al. Computer-Aided multiview tumor detection for automated whole breast ultrasound. Ultrasonic Imaging 2014;36:3-17. [Crossref] [PubMed]
- Zanotel M, Bednarova I, Londero V, et al. Automated breast ultrasound: Basic principles and emerging clinical applications. Radiol Med 2018;123:1-12. [Crossref] [PubMed]
- Zheng FY, Lu Q, Huang BJ, et al. Imaging features of automated breast volume scanner: Correlation with molecular subtypes of breast cancer. Eur J Radiol 2017;86:267-75. [Crossref] [PubMed]
- Grubstein A, Rapson Y, Gadiel I, et al. Analysis of false-negative readings of automated breast ultrasound studies. J Clin Ultrasound 2017;45:245-51. [Crossref] [PubMed]
- Arleo EK, Saleh M, Ionescu D, et al. Recall rate of screening ultrasound with automated breast volumetric scanning (ABVS) in women with dense breasts: A first quarter experience. Clin Imaging 2014;38:439-44. [Crossref] [PubMed]
- Honda E, Nakayama R, Koyama H, et al. Computer-Aided diagnosis scheme for distinguishing between benign and malignant masses in breast DCE-MRI. J Digit Imaging 2016;29:388-93. [Crossref] [PubMed]
- van Zelst J, Tan T, Clauser P, et al. Dedicated computer-aided detection software for automated 3D breast ultrasound;An efficient tool for the radiologist in supplemental screening of women with dense breasts. Eur Radiol 2018;28:2996-3006. [Crossref] [PubMed]
- Tan T, Platel B, Mus R, et al. Computer-aided detection of cancer in automated 3-D breast ultrasound. IEEE Trans Med Imaging 2013;32:1698-706. [Crossref] [PubMed]
- van Zelst JCM, Tan T, Platel B, et al. Improved cancer detection in automated breast ultrasound by radiologists using Computer Aided Detection. Eur J Radiol 2017;89:54-9. [Crossref] [PubMed]
- Moon WK, Lo CM, Chen RT, et al. Tumor detection in automated breast ultrasound images using quantitative tissue clustering. Med Phys 2014;41:42901. [Crossref] [PubMed]
- Kuo HC, Giger ML, Reiser I, et al. Segmentation of breast masses on dedicated breast computed tomography and three-imensional breast ultrasound images. J Med Imaging 2014;1:014501. [Crossref] [PubMed]
- Lo C, Chen R, Chang Y, et al. Multi-dimensional tumor detection in automated whole breast ultrasound using topographic watershed. IEEE Trans Med Imaging 2014;33:1503-11. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. International MICCAI 2015: Medical Image Computing and Computer-Assisted Intervention – MICCAI. 2015:234-41.
- Khened M, Kollerathu VA, Krishnamurthi G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med Image Anal 2019;51:21-45. [Crossref] [PubMed]
- Seely JM, Alhassan T. Screening for breast cancer in 2018-what should we be doing today? Curr Oncol 2018;25:S115-24. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Liao F, Liang M, Li Z, et al. Evaluate the malignancy of pulmonary nodules using the 3-D deep leaky noisy-or network. IEEE Trans Neural Netw Learn Syst 2019;30:3484-95. [Crossref] [PubMed]
- Woo S, Park J, Lee J, et al. CBAM: Convolutional block attention module. ECCV 2018: Computer Vision – ECCV 2018:3-19.
- Liu R, Lehman J, Molino P, Such F P, Frank E, Sergeev A, Yosinski J. An intriguing failing of convolutional neural networks and the coordconv solution. Advances in Neural Information Processing Systems. 2018.
- Huang YL, Chen DR. Watershed segmentation for breast tumor in 2-D sonography. Ultrasound Med Biol 2004;30:625-32. [Crossref] [PubMed]
- Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:640-51. [Crossref] [PubMed]
- Milletari F, Navab N, Ahmadi S. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV). 2016:565-71.
- Zhao H, Shi J, Qi X, et al. Pyramid scene parsing network. 017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI. 2017:6230-9.
- Kozegar E, Soryani M, Behnam H, et al. Computer aided detection in automated 3-D breast ultrasound images: A survey. Artif Intell Rev 2020;53:1919-41. [Crossref]
- Omidiji OA, Campbell PC, Irurhe NK, et al. Breast cancer screening in a resource poor country: Ultrasound versus mammography. Ghana Med J 2017;51:6-12. [Crossref] [PubMed]
- Pesapane F, Codari M, Sardanelli F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp 2018;2:35. [Crossref] [PubMed]
- Wang S, Wang R, Zhang S, et al. 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤ 3 cm using HRCT. Quant Imaging Med Surg 2018;8:491-9. [Crossref] [PubMed]
- Mazurowski MA, Buda M, Saha A, et al. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging 2019;49:939-54. [Crossref] [PubMed]
- Wu C, Guo S, Hong Y, et al. Discrimination and conversion prediction of mild cognitive impairment using convolutional neural networks. Quant Imaging Med Surg 2018;8:992-1003. [Crossref] [PubMed]
- Sun D, Wang M, Li A. A multimodal deep neural network for human breast cancer prognosis prediction by integrating Multi-Dimensional data. IEEE/ACM Trans Comput Biol Bioinform 2018;16:841-50.
- Yap MH, Pons G, Martí J, et al. Automated breast ultrasound nodule detection using convolutional neural networks. IEEE J Biomed Health Inform 2018;22:1218-26. [Crossref] [PubMed]
- Li J, Sarma KV, Chung HK, et al. A multi-scale U-Net for semantic segmentation of histological images from radical prostatectomies. AMIA Annu Symp Proc 2018;2017:1140-48. [PubMed]
- Wang S, Zhang R, Deng Y, et al. Discrimination of smoking status by MRI based on deep learning method. Quant Imaging Med Surg 2018;8:1113-20. [Crossref] [PubMed]
- Xu Y, Wang Y, Yuan J, et al. Medical breast ultrasound image segmentation by machine learning. Ultrasonics 2019;91:1-9. [Crossref] [PubMed]
- Çiçek Ö, Abdulkadir A, Lienkamp SS, et al. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. Medical Image Computing and Computer-Assisted Intervention. 2016.
- Huang Q, Zhang F, Li X. Machine learning in ultrasound Computer-Aided diagnostic systems: A survey. Biomed Res Int 2018;2018:5137904. [Crossref] [PubMed]
- Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, et al. UNet++: A nested U-Net architecture for medical image segmentation. Cham: Springer International Publishing, 2018.
- Kumar V, Webb JM, Gregory A, et al. Automated and real-time segmentation of suspicious breast masses using convolutional neural network. PLoS One 2018;13:e0195816. [Crossref] [PubMed]
- Pons G, Martí J, Martí R, et al. Breast-lesion Segmentation Combining B-Mode and Elastography Ultrasound. Ultrason Imaging 2016;38:209-24. [Crossref] [PubMed]
- Chiang TC, Huang YS, Chen RT, et al. Tumor detection in automated breast ultrasound using 3-D CNN and prioritized candidate aggregation. IEEE Trans Med Imaging 2019;38:240-49. [Crossref] [PubMed]
- Moon WK, Shen YW, Bae MS, et al. Computer-aided tumor detection based on multi-scale blob detection algorithm in automated breast ultrasound images. IEEE Trans Med Imaging 2013;32:1191-200. [Crossref] [PubMed]
- Guo Y, Hu Y, Qiao M, et al. Radiomics analysis on ultrasound for prediction of biologic behavior in breast invasive ductal carcinoma. Clin Breast Cancer 2018;18:e335-44. [Crossref] [PubMed]