P2RY14 downregulation in lung adenocarcinoma: a potential therapeutic target associated with immune infiltration
Introduction
Lung cancer is the most common cause of cancer-related death worldwide, with approximately 2.1 million new lung cancer cases and an estimated 1.8 million fatalities occurring in 2018 (1). Non-small cell lung cancer (NSCLC) is responsible for 80–85% of lung cancers based on histological characteristics (2), while lung adenocarcinoma (LUAD) is the most frequent pathological type of NSCLC, accounting for roughly 50% of cases (3). Despite significant advancements in early detection, targeted therapy, and immunotherapy, most patients suffering from LUAD are detected at an advanced stage due to a lack of early clinical signs and adequate diagnostic tools (4). The prognosis for people with operable lung cancer is likewise considerably variable. For example, patients at stage Ia following surgery had a 5-year overall survival (OS) of 73%, while those at stage IIIa had a 5-year OS of only 24% (5). As a result, it is critical to identify patients suffering from LUAD who have poor prognostic characteristics and administer more active treatments to maximize therapeutic benefits. Despite numerous clinicopathological indicators that can potentially predict the prognosis of patients suffering from LUAD such as tumor size, lymph node metastases, and positive margin, prognosis and recurrence are still uneven in patients with the same stage and treatment. As a result, additional simple and effective biomarkers for predicting patient prognosis and recurrence are needed.
The TRACERx lung cancer research program in the United Kingdom shows that the early “evolution” of LUAD is mainly due to mutations or amplifications such as EGFR, MET, BRAF and TP53, which mainly occur at the molecular clone level. These molecules are related to the formation of NSCLC and belong to driver genes. But late-stage studies of the “evolution” of NSCLC adenocarcinomas found that 75% of tumors had various variants at the subclonal level, which could be located in different regions of the tumor, resulting in intratumor heterogeneity (6-9). However, these heterogeneous LUADs with different molecular types have different immune microenvironments, and also have different responses to the same immunosuppressive agents, resulting in different prognosis. Therefore, by finding genes with different immune signatures, we can better guide the treatment of LUAD patients with different molecular types and improve the prognosis of these patients.
A member of the original family of extracellular nucleotide sensing receptors, purinergic (P2) receptors appeared at an early stage in evolution (10). Fifteen subtypes have been cloned to date and are roughly divided into G protein-coupled P2Y receptors (P2Y 1, 2, 4, 6, 11–14) and ionic P2X receptors (P2X 1–7). The P2Y14 receptor is the only member of the P2 receptor family that responds specifically to UDP-sugars, such as UDP-glucose (UDPG), and UDP-galactose (11). The extracellular function of UDP-sugar is that it can be released into the extracellular space through Ca2+-regulated exocytosis, where it has enzyme stability and can stimulate P2Y14 receptors (12-14). P2Y14 has been linked to the onset and development of malignancies and has been found to play a role in the immune system by contributing to the control of stem cell compartments (15). The activated P2Y14 receptor suppresses glioma cell growth and development by lowering interleukin-6 (IL-6) production (16). In the formation and development of gastrointestinal cancers, P2Y14 receptor-mediated signal cascades have a regulatory role in intracellular ion concentration and trigger numerous mitogen-activated protein kinases (MAPKs), Src family kinases, and downstream protein kinases (17).
To find differentially expressed genes (DEGs) linked to lung cancer prognosis, we employed the intersection of gene datasets related to LUAD and immune infiltration-related datasets from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. The usefulness of this gene was further addressed using clinical data from patients suffering from LUAD at our hospital to obtain a more realistic clinical reference, which is valuable for the timely screening of patients with poor prognostic features. And we lay the foundation for future basic research through further molecular mechanism prediction. From the perspective of database-clinical-basic, it is explained that the expression of this molecule is significantly related to immune infiltration, and it may play an important inhibitory role in the immune escape of tumor cells in the LUAD microenvironment. It is helpful for the development of targeted precision oncology and has good innovation and reference value. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-115/rc).
Methods
Screening of DEGs
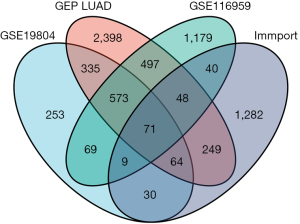
The GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) was employed to download RNA expression data sets GSE19804 and GSE116959 (contains normal tissue and tumor tissue) through the GEOquery package, and the probe corresponding to multiple molecules was removed, retaining only the probe with the largest signal value when encountered with multiple probes corresponding to the same molecule. The dataset of DEGs in LUAD samples was obtained from TCGA database (https://tcga- data.nci.nih.gov/). From the gene list module of the Immunology Database and Analysis Portal (ImmPort) database [ImmPort Private Data (nih.gov)], we directly downloaded the complete gene names, including a total of 2,483 immune-associated genes (uploaded as an attachment). By taking the intersection of these four datasets and making a Venn diagram, with |log2FC| >1 and P<0.05 as the parameters to determine the screening of DEGs, we obtained the P2RY14 gene, which is associated with the long-term outcome of LUAD and related to immunity.
Verification of P2RY14 in TCGA database
We analyzed LUAD tissues and para-cancer tissues from unpaired samples and paired samples in TCGA database (https://tcga-data.nci.nih.gov/) and compared the expression of DEG P2RY14 in both samples.
Expression of P2RY14 at the organizational level in the Human Protein Atlas (HPA) database
We clarified the location of P2RY14 by querying its expression in the HPA database (https://www.proteinatlas.org/search/HAMP).
Relationship between P2RY14 and clinical data of LUAD in TCGA database
The correlation between P2RY14 and the prognosis of patients with LUAD and their correlation with clinical data was ultimately carried out by analyzing the clinical data of LUAD in TCGA database.
Relationship between P2RY14 and clinical data of LUAD in the Kaplan-Meier (KM)-plotter database
We further validated the accuracy of the data following the analysis of the clinical data of LUAD in the KM-plotter database (https://kmplot.com/analysis/index.php?p=service), constructing forest plots to further analyze the correlation of P2RY14 and clinical data, and comparing with the results in the TCGA database.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
Using Metascape online function analysis (https://metascape.org/gp/index.html#/main/step1), the differential gene was added to Metascape for functional analysis.
Analysis of genes interacting with P2RY14 in the GeneMANIA database and string database
We analyzed the related genes interacting with P2RY14 from the perspective of protein-DNA, protein-protein, genetic interaction pathway, gene and protein expression, biochemical and physiological reactions, phenotype screening, and protein domain via the GeneMANIA database (https://genemania.org/) and string database (https://string-db.org/) and generated a protein-protein interaction (PPI) network.
Correlation between P2RY14 and immune cells in the Tumor Immune Estimation Resource (TIMER) database
We constructed a lollipop chart through the TIMER database (https://cistrome.shinyapps.io/timer/) to show the correlation between P2RY14 expression in LUAD and tumor purity and immune cells.
Correlation between P2RY14 and immune checkpoint related molecules in Gene Expression Profiling Interactive Analysis (GEPIA) database
Analysis of the correlation between three immune checkpoints (CD274, PDCD1, CTLA4) and P2RY14 was made using the GEPIA database (http://gepia.cancer-pku.cn/index.html).
Correlation between P2RY14 and immune-related expressed genes analyzed in the TIMER database
The correlation of P2RY14 with immune-related expressed genes and the correlation with genes of different types of T cells were analyzed in the TIMER database (https://cistrome.shinyapps.io/timer/). At the same time, a forest map and KM survival curve were drawn to further analyze the different expression of P2RY14 in different immune cells and the prognosis of LUAD.
Selection of clinical data
The clinical data of 100 patients undergoing pulmonary adenocarcinoma surgery admitted to Nanjing Chest Hospital from January 2013 to June 2016 was selected. Inclusion criteria was as follows: (I) patients who were diagnosed with LUAD for the first time and had to undergo surgery; (II) postoperative histopathology confirmed LUAD; (III) complete clinical and follow-up data; (IV) no other anti-tumor surgery treatment prior to hospital admission; (V) good compliance. Exclusion criteria: (I) patients with positive pathological resection margins after operation; (II) combined with other serious diseases, such as severe diabetes, heart failure, or chronic obstructive pulmonary disease; (III) complications of a serious nature occurring in the perioperative period; (IV) patients who refused follow-up. In total, 100 patients were enrolled including 67 males and 33 females, whose age ranged from 36 to 79 years, with a mean age of 62.3±8.4 years.
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Nanjing Chest Hospital (No. 2021-KY094-01) and informed consent was taken from all the patients.
Tissue microarray construction and immunohistochemistry (IHC)
Tissue samples were obtained from patients who experienced lung cancer surgery, and the Pathology Department at Nanjing Chest Hospital created the tissue microarray. Hematoxylin and eosin-stained paraffin blocks of the 100 cases of LUAD were obtained and the most common features were selected and labeled at fixed positions under a microscope, and less than 160 dots in each dot array were observed. A 3 m slice of the receptor block was cut and shifted to a glass slide with the help of a tape transfer technique for UV cross-linking. ab140896 was used as the antibody for P2RY14, and during immunohistochemical evaluation, the intensity of cell staining and the proportion of positive cells were used to rate the outcome. The staining intensity was assessed as follows: 0 point (negative), 1 point (25%), 2 points (25–50%), 3 points (51–75%), and 4 points (>75%), and as 0 point (negative or no staining), 1 point (weakly positive), 2 points (moderately positive), 3 points (moderately positive), 4 points (>75%) (strongly positive). The final score for the individual specimen was computed by multiplying the two scores together. After taking the arithmetic mean of the scores, specimens with a score less than 7 were labeled as having low P2RY14 expression.
Quantitative reverse transcription PCR (RT-qPCR) method to detect mRNA encoding P2RY14
Tumor tissue and nearby normal tissue samples were obtained during the operation and cryopreserved in liquid nitrogen tanks. We then ground 100 mg tumor tissue and nearby normal tissue samples into powder using the liquid nitrogen milling process, 1 mL Trizol lysis solution was added, and total RNA was extracted as directed. The primers were: forward 5'-TTCTGGGTCGTGTTTCTTCTG-3' and reverse 5'-CGAGAGTAGCAGAGTGAATTC-3'. GAPDH primers (370 bp product) were: forward 5'-CTCATGACCACAGTCCATGC-3' and reverse 5'-GGTCCAGGGGTCTTACTCC-3'. The conditions employed during PCR for P2RY14 were 94 °C for 1 minute, 50 °C for 1.5 minutes, and 72 °C for 2 minutes, with 40 cycles. The PCR conditions for GAPDH were 94 °C for 1 minute, 58 °C for 1 minute, and 72 °C for 1 minute, with 30 cycles. The PCR products were electrophoresed on a 1% (wv-1) agarose gel and stained with ethidium bromide. To rule out the possibility of genomic DNA contamination, GAPDH primers were used to run PCR experiments prior to cDNA synthesis. Subsequently, the 2−ΔΔCt method was employed to estimate the mRNA relative expression of the target molecule for clarifying the expression in cancer and para-cancer tissue.
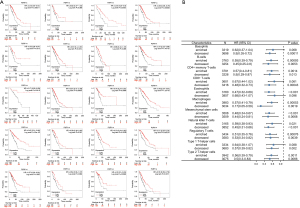
Correlation between P2RY14 and the survival prognosis and clinical data parameters of patients with LUAD
The study categorized 100 patients with LUAD into two groups based on P2RY14 IHC expression: a P2RY14 high-expression group and P2RY14 low-expression group. Association between the data of the groups and the survival prognosis and clinical parameters of the patients was compared using SPSS software. Simultaneously, univariate and multivariate analyses were employed to screen the independent prognostic markers that are relevant for the prognosis of individuals suffering from lung cancer.
Construction of nomogram
The independent factors affecting the prognosis of patients suffering from LUAD were obtained by Cox regression analysis, R language was edited, and the nomogram was constructed.
Follow-up
Outpatient review, re-admission medical records, telephone follow-up, and other means of follow-up were employed. Patients were followed up once every 3 months in the first year, once every 6 months in the second year, and once each year after 2 years. The end of follow-up was either the follow-up deadline or death of the patient. The follow-up deadline was July 1, 2021, and the survival time was referred to as the time between the start of follow-up and death or the mentioned deadline. The OS was defined as the duration from the diagnosis of the disease to the end of the follow-up period or death from any cause.
Statistical analysis
For statistical analysis and visualization, R (version 3.6.3) was utilized. For data download, DESeq2 (version 1.26.0) R package was used and for difference analysis, Limma package (version 3.42.2) was employed. For statistical analysis, SPSS 22.0 (IBM, Armonk, NY, USA) software was used. The clinicopathological conditions of the two groups of patients were compared using the chi-square test, the KM approach was employed to assess patient survival, and the log-rank statistical method was conducted to test significance. Finally, the Cox proportional risk regression model was utilized to find the relevant independent prognostic markers for bladder cancer (BLCA) patients’ prognosis. Eventually, a nomogram was drawn using the R programming language, with a statistical significance of P<0.05.
Results
Selection of DEGs
The intersection of two RNA expression data sets; GSE19804 and GSE116959 was obtained from the GEO database, the data set of DEGs of LUAD from TCGA database, and gene list from the ImmPort database. A Venn diagram was then constructed, and using |log2FC| >1 and P<0.05 as the parameters to determine the screening of DEGs, the P2RY14 gene which is linked with the prognosis of LUAD and related to immunity was obtained (Figure 1).
P2RY14 expression of in TCGA database
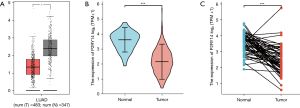
We referred to TCGA database for an analysis of the data of LUAD tissue and para-cancer tissues and compared the two in terms of the expression of the DEGs P2RY14. We concluded that in both groups, para-cancer tissues had an elevated P2RY14 expression in comparison to cancer tissues in unpaired LUAD tissue specimens (Figure 2A: 483 cases of cancer tissues, 347 cases of para-cancer tissues; Figure 2B: 535 cases of cancer tissues, 59 cases of para-cancer tissues). In paired LUAD tissue samples, para-cancer tissues had a higher P2RY14 expression in comparison to cancer tissues (Figure 2C: 57 cases of cancer tissues, 57 cases of para-cancer tissues).
P2RY14 expression at tissue level in the HPA database
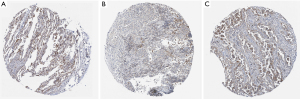
By querying the expression of P2RY14 in the HPA database, we found P2RY14 was primarily located within the cell cytoplasm (Figure 3A-3C). Immunohistochemical results showed that para-cancer tissues had a higher P2RY14 expression in comparison to cancer tissues. In addition, P2RY14 expression in cancer tissues was higher in the case of well-differentiated patients (pathological differentiation of specimens in Figure 3C was better than that in Figure 3B).
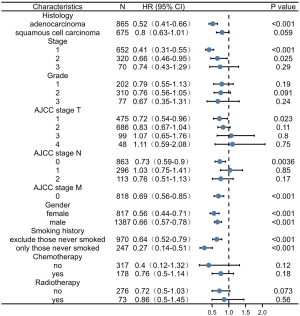
Relationship between P2RY14 and clinical data of LUAD in TCGA database
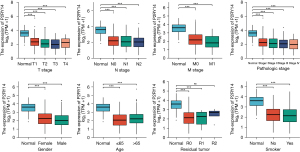
By examining TCGA database based clinical data pertaining to LUAD, we clarified the link between the P2RY14 molecule and prognosis of LUAD patients (Figure 4A-4C). We found individuals suffering from LUAD with a high P2RY14 expression in cancer tissues had better OS and disease-specific survival (DSS) in comparison to those with a low P2RY14 expression. In further comparison with clinical data, the expression of P2RY14 was found to be different in LUAD patients with different TNM stages, different pathological stages, different genders, different ages, those with or without residual tumor tissue, and those with or without a smoking history. These results confirmed patients with a high P2RY14 expression have better prognostic factors (Figure 5).
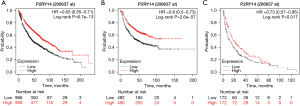
Interrelation between P2RY14 and clinical data of LUAD in the KM-plotter database
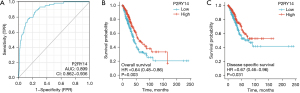
We also investigated the clinical data of LUAD in the KM-plotter database to observe the prognosis of patients with different P2RY14 expression. We discovered patients having a high P2RY14 expression had a better OS (Figure 6A), progression-free survival (PFS) (Figure 6B), and post-progression survival (PPS) (Figure 6C) in comparison to those with a low P2RY14 expression, which corroborated with the expression results in TCGA database above. In addition, a detailed analysis of the correlation between clinical data and P2RY14 was carried out by constructing a forest map (Figure 7), further signifying its clinical implications in LUAD.
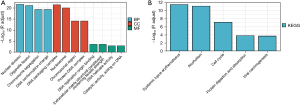
GO and KEGG enrichment analysis
Metascape was employed for conducting online functional analysis. DEGs were added to Metascape for functional analysis of GO (Figure 8A) and KEGG (Figure 8B), respectively, to predict the signaling pathway that P2RY14 may be involved in. We found that P2RY14 may affect the biological events of LUAD by participating in the origin of DNA replication, cell cycle, chromosome separation, and DNA conformation change, leading to different prognoses of LUAD, which also provides a reference for the specific mechanism of action in future basic experimental studies.
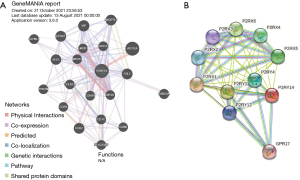
Analysis of genes interacting with P2RY14 in GeneMANIA database and string database
We analyzed the related genes interacting with P2RY14 from the perspective of protein-DNA, protein-protein and genetic interaction pathway, gene and protein expression, biochemical and physiological reactions, protein domain, and phenotype screening through the string database (https://string-db.org/) and GeneMANIA database (https://genemania.org/), and constructed a PPI network. The top 20 genes were screened from the two databases according to the intensity of the interaction (Figure 9).
Correlation between P2RY14 and immune cells in TIMER database
Considering the poor prognosis of LUAD and the use of immunosuppressive agents in clinical treatment, we obtained the P2RY14 molecule from the intersection of DEGs of LUAD and immune-related genes. We then used the TIMER database to construct a lollipop chart to show the link between P2RY14 expression and tumors purity and each immune cell in LUAD. P2RY14 expression was found to be positively correlated with six kinds of infiltrating immune cells, including B cells, dendritic cells (DCs), neutrophils, macrophages, CD4+ T cells, and CD8+ T cells (Figure 10A). To identify the impact of P2RY14 on the tumor microenvironment (TME) in further detail, we analyzed the correlation between specific immune cells and P2RY14, and the outcome revealed that P2RY14 had a positive correlation with the extent of infiltration of DCs, macrophages, neutrophils, T cells, and Th1 cells, while P2RY14 had a negative correlation with the extent infiltration levels of CD56birghtNK cells and Th2 cells (Figure 10B). Additional analysis revealed P2RY14 expression had a positive correlation with the immune checkpoint related molecules CD274, CTLA-4, and PDCD1 (Figure 10C). The considerable correlation between the expression of P2RY14 and immune infiltration is further supported by these findings and implies that P2RY14 may have an indispensable role in inhibiting immune escape of tumor cells in the microenvironment of LUAD, which also provides a better reference for future basic research.
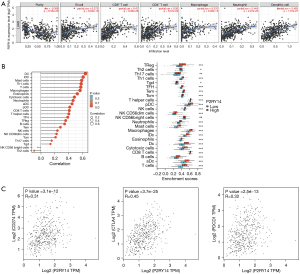
Correlation analysis between P2RY14 and immune-related expressed genes in the TIMER database
To further understand details of the interaction among P2RY14 and the immune response, we carried out a TIMER based analysis of the correlation between P2RY14 and immune-related expressed genes. Table 1 lists the genes utilized for the characterization of immune cells, and includes T cells, B cells and CD8+ T cells, tumor-associated macrophages (TAM), monocytes, DCs, neutrophils, natural killer (NK) cells, and M1and M2 macrophages. Tumor purity is a significant factor influencing the separation of immune infiltration in clinical tumor biopsy, and following adjustment for it, hepcidin expression was found to have a significant association with most of the immune markers in different types of immune cells in LUAD and lung squamous cell carcinoma (LUSC) (Table 1). The link between P2RY14 expression and many functional T cells, including Treg, resting Treg, Th1, Th1-like, and Th2, was also detected. We discovered with the aid of the TIMER database, that P2RY14 expression levels had a considerable link with 11 of the 12 T cell markers in LUAD following adjustment for tumor purity (Table 2). The earlier findings depicted that P2RY14 was related to the prognosis of LUAD and immune infiltration. For the purpose of further investigation of the effect of P2RY14 on the prognosis of LUAD caused by immune infiltration, the KM-plotter database was again used for in-depth analysis of the tumor survival rate under the condition of the presence or absence of immune cells (Figure 11A). Finally, these data were made into a forest map (Figure 11B) to reflect the results more intuitively and showed patients with LUAD with a low expression of P2RY14 and reduced infiltration of CD4+ memory T cells, B cells, macrophages, and NK cells had a poor prognosis. The findings again suggest P2RY14 can potentially impact the prognosis of patients with LUAD through immune infiltration.
Table 1
| Description | Gene markers | BLCA | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| Cor | P | Cor | P | |||
| B cell | CD19 | 0.3276 | *** | −0.4921 | *** | |
| CD79A | 0.3136 | *** | −0.4571 | *** | ||
| T cell (general) | CD3D | 0.5179 | *** | −0.5169 | *** | |
| CD3E | 0.5551 | *** | −0.5361 | *** | ||
| CD2 | 0.5885 | *** | −0.5218 | *** | ||
| CD8+ T cell | CD8A | 0.4636 | *** | −0.4368 | *** | |
| CD8B | 0.3909 | *** | −0.3483 | *** | ||
| Monocyte | CD86 | 0.5772 | *** | −0.4499 | *** | |
| CSF1R | 0.5210 | *** | −0.3956 | *** | ||
| TAM | CCL2 | 0.3380 | *** | −0.3336 | *** | |
| CD68 | 0.4372 | *** | −0.3590 | *** | ||
| IL10 | 0.5050 | *** | −0.4196 | *** | ||
| M1 | IRF5 | 0.2355 | *** | −0.3368 | *** | |
| PTGS2 | −0.0196 | 0.6573 | −0.0190 | 0.673920 | ||
| NOS2 | 0.2971 | *** | −0.2295 | *** | ||
| M2 | CD163 | 0.4734 | *** | −0.3846 | *** | |
| VSIG4 | 0.4709 | *** | −0.3295 | *** | ||
| MS4A4A | 0.5810 | *** | −0.4088 | *** | ||
| Neutrophils | CEACAM8 | 0.2990 | *** | −0.0660 | 0.142980 | |
| ITGAM | 0.4453 | *** | −0.3632 | *** | ||
| CCR7 | 0.5681 | *** | −0.5215 | *** | ||
| NK cell | KIR2DL1 | 0.1913 | *** | −0.1527 | *** | |
| DC | HLA-DPB1 | 0.5604 | *** | −0.3880 | *** | |
***P<0.001. TIMER, Tumor Immune Estimation Resource; BLCA, bladder cancer; TAM, tumor-associated macrophages; NK, natural killer; DC, dendritic cell.
Table 2
| Description | Gene markers | LUAD | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| Cor | P | Cor | P | |||
| Th1 | TBX21 | 0.4171 | *** | −0.4521 | *** | |
| STAT4 | 0.4450 | *** | −0.4563 | *** | ||
| STAT1 | 0.2806 | *** | −0.3285 | *** | ||
| TNF | 0.3715 | *** | −0.3976 | *** | ||
| IFNG | 0.2649 | *** | −0.3493 | *** | ||
| Th1-like | CXCR3 | 0.3892 | *** | −0.4319 | *** | |
| BHLHE40 | 0.0522 | 0.2367 | −0.1059 | * | ||
| CD4 | 0.6495 | *** | −0.4779 | *** | ||
| Th2 | STAT6 | 0.1991 | *** | 0.0262 | 0.5614 | |
| STAT5A | 0.4570 | *** | −0.4142 | *** | ||
| Treg | FOXP3 | 0.4639 | *** | −0.4758 | *** | |
| Resting Treg | IL2RA | 0.5257 | *** | −0.3877 | *** | |
*P<0.05; ***P<0.001. TIMER, Tumor Immune Estimation Resource; LUAD, lung adenocarcinoma.
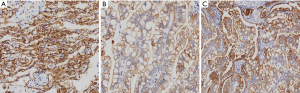
Expression of P2RY14 in the IHC of the specimens of patients
While the results from immunohistochemical analysis suggested both tumor and para-cancer tissues of LUAD showed P2RY14 expression, the latter had a higher P2RY14 expression in comparison to the former tumor (Figure 12).
P2RY14 mRNA expression in LUAD tissue
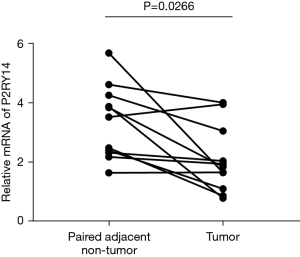
The tumor tissues of 11 patients included in the study were detected, and the para-cancer tissues were used as the control. RT-qPCR was employed to detect the mRNA encoding P2RY14 and showed it was expressed in both para-cancer tissues and LUAD tumor tissues and was higher in the former (Figure 13).
Correlation between P2RY14 and the survival prognosis and clinical data parameters of patients with LUAD
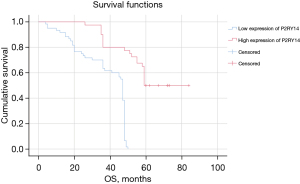
As suggested by the P2RY14 expression revealed in immunohistochemistrical analysis, patients in the study were categorized into two groups: a P2RY14 high-expression group and a P2RY14 low-expression group. SPSS software was used to draw a comparison of the correlations among the two groups of data and the survival prognosis and clinical parameters of the patients. At the same time, the independent prognostic factors of any significance for the prognosis of individuals suffering from LUAD were screened by univariate and multivariate analysis. The final study comprised a total of 100 patients, of which there were 80 deaths and 20 surviving patients revealing a mortality rate and survival rate of 80.0% and 20.0%, respectively, and a median OS of 48 months. The P2RY14 high-expression group had a considerably enhanced OS in comparison to that of the P2RY14 low-expression group (P<0.05) (Figure 14). Furthermore, we found considerable variations in pathological staging and differentiation degree between the P2RY14 high and low expression groups (P<0.05) (Table 3), and Cox regression analysis revealed that for the prognosis of LUAD patients, P2RY14 expression, the degree of differentiation, and smoking history were independent risk factors (P=0.022; P=0.044; P=0.000) (Tables 4,5).
Table 3
| Variables | Number of cases (n=100) |
P2RY14 low-expression (n=60) | P2RY14 high-expression (n=40) | χ2 | P |
|---|---|---|---|---|---|
| Gender | 0.611 | 0.435 | |||
| Female | 33 | 18 | 15 | ||
| Male | 67 | 42 | 25 | ||
| Age (years) | 1.939 | 0.164 | |||
| ≤65 | 54 | 29 | 25 | ||
| >65 | 46 | 31 | 15 | ||
| Tumor diameter (cm) | 1.752 | 0.186 | |||
| ≤3 | 42 | 22 | 20 | ||
| >3 | 58 | 38 | 20 | ||
| Pathological staging | 9.722 | 0.002* | |||
| Stage 1 + stage 2 | 70 | 35 | 35 | ||
| Stage 3 + stage 4 | 30 | 25 | 5 | ||
| Differentiation degree | 15.366 | 0.000* | |||
| Moderate to high differentiation | 51 | 21 | 30 | ||
| Poor differentiation | 49 | 39 | 10 | ||
| Lymph node metastasis | 0.733 | 0.392 | |||
| None | 65 | 37 | 28 | ||
| Yes | 35 | 23 | 12 | ||
| Smoking history | 1.166 | 0.280 | |||
| None | 29 | 15 | 14 | ||
| Yes | 71 | 45 | 26 | ||
| Drinking history | 0.008 | 0.927 | |||
| None | 73 | 44 | 29 | ||
| Yes | 27 | 16 | 11 | ||
| Diabetes | 2.217 | 0.145 | |||
| None | 96 | 59 | 37 | ||
| Yes | 4 | 1 | 3 | ||
| Hypertension | 1.175 | 0.278 | |||
| None | 78 | 49 | 29 | ||
| Yes | 22 | 11 | 11 |
*P<0.05 shows that the variation was statistically considerable.
Table 4
| Variables | HR | 95% CI | P |
|---|---|---|---|
| Gender | |||
| Female | 1.309 | 0.815–2.101 | 0.265 |
| Male | 1 | ||
| Age (years) | |||
| ≤65 | 1.322 | 0.849–2.058 | 0.217 |
| >65 | 1 | ||
| Tumor diameter (cm) | |||
| ≤3 | 1.006 | 0.646–1.568 | 0.977 |
| >3 | 1 | ||
| Pathological staging | |||
| Stage 1 + stage 2 | 0.543 | 0.339–0.868 | 0.011* |
| Stage 3 + stage 4 | 1 | ||
| Differentiation degree | |||
| Moderate to high differentiation | 0.358 | 0.228–0.562 | 0.000* |
| Poor differentiation | 1 | ||
| Lymph node metastasis | |||
| None | 0.871 | 0.550–1.379 | 0.556 |
| Yes | 1 | ||
| Smoking history | |||
| None | 0.418 | 0.241–0.726 | 0.002* |
| Yes | 1 | ||
| Drinking history | |||
| None | 0.977 | 0.598–1.597 | 0.926 |
| Yes | 1 | ||
| Diabetes | |||
| None | 0.999 | 0.315–3.171 | 0.999 |
| Yes | 1 | ||
| Hypertension | |||
| None | 1.399 | 0.818–2.392 | 0.220 |
| Yes | 1 | ||
| P2RY14 expression | |||
| Low-expression | 8.693 | 4.139–18.258 | 0.000* |
| High-expression | 1 |
*P<0.05 shows that the variation was statistically considerable. OS, overall survival; HR, hazard ratio; CI, confidence interval.
Table 5
| Variables | HR | 95% CI | P |
|---|---|---|---|
| Differentiation degree | |||
| Moderate to high differentiation | 1.739 | 1.081–2.797 | 0.022* |
| Poor differentiation | 1 | ||
| Smoking history | |||
| None | 1.791 | 1.017–3.154 | 0.044* |
| Yes | 1 | ||
| P2RY14 expression | |||
| Low-expression | 0.148 | 0.070–0.316 | 0.000* |
| High-expression | 1 |
*P<0.05 implies that the variation was statistically considerable. OS, overall survival; HR, hazard ratio; CI, confidence interval.
Nonogram construction
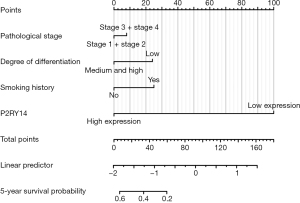
The independent factors that influence the prognosis of patients suffering from LUAD were analyzed by Cox regression method. The R language was then edited, and a nomogram was constructed. The nomogram C-index was 0.774 [95% confidence interval (CI): 0.744–0.804] as suggested by independent verification (Figure 15), showing a certain accuracy in predicting the 5-year survival rate of individuals suffering from LUAD. It also had certain guiding significance for screening patients with poor prognostic characteristics in clinical work and improving the prognosis of these patients.
Discussion
Lung cancer is the most common malignant tumor and is the main underlying cause of cancer-associated death in both men and women worldwide (1). Despite clinical advances in early diagnosis, targeted treatment, and immunotherapy, lung cancer is frequently diagnosed at a progressive stage, with a poor prognosis (2,18), including the most common pathological type of NSCLC, LUAD (19,20). As a result, it is critical to investigate the mechanisms that lead to LUAD metastasis and the identification of useful prognostic biomarkers for the disease. Immunotherapy techniques based on immune microenvironment modulation are now being used in clinical practice and have become a hot topic in the treatment of LUAD (21,22). The FDA has also approved a number of therapeutic antibodies for the treatment of LUAD, but numerous issues remain concerning the best dosage and timing for medications that block these immune checkpoint channels.
The genes included in the molecular typing of LUAD often include EGFR, ALK, ROS1, BRAF, NTRK1/2/3, MET, and RET, and the mutation rate of functional driver genes is about 60%, among which KRAS, EGFR mutations and EML4-ALK fusion is the most common driver gene, accounting for about 35% to 40% (23-25). The study by Jones et al showed that KRAS G12C mutation was associated with worse disease-free survival after complete resection of stage I–III LUAD (26).
In the current work, we first downloaded data related to the prognosis of LUAD from the GEO and TCGA databases and took the intersection with the immune-related data set in ImmPort database to screen out the DEG P2RY14, which was related to immune infiltration as well as prognosis of LUAD. We then authenticated the expression of P2RY14 in TCGA database and found that as it was more highly expressed para-cancer compared to cancer tissues, the expression of this molecule was more similar to a “tumor suppressor gene”. Subsequently, we observed through the HPA database, that P2RY14 primarily resides in the cell cytoplasm. Interestingly, in para-cancer tissues, the expression of P2RY14 was comparatively higher in comparison to cancer tissues, and its expression in cancer tissues was found to be elevated in well-differentiated patients, which is in agreement with the expression in TCGA database and stimulated our interest in further research. We then analyzed the interrelation among the P2RY14 molecule and the prognosis of individuals suffering from LUAD in the TCGA database and found patients with a high P2RY14 expression in LUAD tissues had a better OS and DSS in comparison to those with low P2RY14 expression. In further comparison with clinical data, it was observed that P2RY14 expression was different in patients suffering from LUAD with different TNM stages, different pathological stages, different genders, different ages, with or without residual tumor tissue, and with or without a smoking history. Patients with high P2RY14 expression also had more factors associated with a better outcome. Patients suffering from LUAD who had high P2RY14 expression had higher OS, PFS, and PPS than those with low expression, as suggested by the KM-plotter database. These findings support the idea that P2RY14 could be a standalone predictive biomarker for lung cancer and could aid in the development of targeted precision oncology.
We also conducted online GO and KEGG function analyses using Metascape, and found that P2RY14 may affect biological events of LUAD by participating in DNA replication origin, cell cycle, chromosome separation, and DNA conformation change, leading to different prognoses of LUAD. This result also provides a reference for the specific mechanism of action in future basic experimental research. Genes interacting with P2RY14 were screened out by the GeneMANIA database and string database, and the top 20 were screened out according to the intensity of action. This revealed these interacting genes were also important in the TME. According to Kim et al. (27), the preferential upregulation of PRX 1 in lung cancer cells may represent their attempt to influence dynamic REDOX alterations, proliferation, and malignant progression in the TME in a way that is beneficial to them. Chua et al. (28) reported PRX3 might be a candidate proliferation marker of breast cancer through its regulation of cell cycle. The interaction of GPR17-T0510-3657 modulated the interaction between the PX domain and proteins containing the spiral mPTS recognition domain to act as therapeutic targets for glioblastoma multiforme (29). Liu et al. suggested that B cells are an independent prognostic factor in patients with LUAD. B cell [CD19, toll-like receptor 10 (TLR10) and Fc-like receptor A (FCRLA)] and myeloid DC (ITGB2, LAPTM5 and SLC7A7) related genes partially elucidate the role of B cell/DC1 in predicting immune checkpoint blockade efficacy (30). TCGA dataset also demonstrates the gene landscape and correlation between B-cell infiltration and programmed death ligand 1 expression in LUAD patients (31). These findings add to our understanding of P2RY14’s likely role in the immune microenvironment. P2RY14 expression was positively correlated with six kinds of infiltrating immune cells, including B cells, CD8+ T cells, CD4+ T cells, DCs, neutrophils, and macrophages according to the TIMER database’s lollipop chart, as well as immune checkpoint related molecules CD274, CTLA-4, and PDCD1. These findings back up the significant link between P2RY14 expression and immune invasion, implying it may play a key role in preventing tumor cells from escaping the immune system in the microenvironment of LUAD, and providing a better reference point for future basic research. To learn more about the link between P2RY14 and the immune response, we utilized the TIMER database to examine the link between P2RY14 and immune-related expression genes, and in LUAD, P2RY14 expression was substantially linked with most immunological markers in various types of immune cells and was also linked to a variety of functional T cells, including Treg, resting Treg, Th1, Th1-like, and Th2. In LUAD, P2RY14 expression was found to be substantially linked with 11 of the 12 T cell markers. To further determine whether the influence of P2RY14 on the prognosis of LUAD was caused by immune infiltration, we used the KM-plotter database again for in-depth analysis of tumor survival rate under the condition of the presence or absence of immune cells. Patients suffering from LUAD with a low P2RY14 expression and reduced infiltration of macrophages, CD4+ memory T cells, B cells, and NK cells had a poor prognosis, again suggesting P2RY14 has the potential to influence the survival rate of individuals suffering from LUAD by influencing immune infiltration.
After exploring P2RY14 in the database, we conducted further research to verify its effects on the patients included in our study. IHC of these tissue samples showed that while P2RY14 was localized in the cytoplasm and was expressed in both para-cancer and cancer tissues of LUAD, para-cancer tissues had a higher expression in comparison to cancer tissues, and the results of RT-qPCR were consistent with those of IHC. The final study comprised a total of 100 patients, in which there were 80 deaths and 20 surviving patients showing a mortality rate and survival rate of 80.0% and 20.0%, respectively, and a median OS of 48 months. Taking into view the immunohistochemical expression of P2RY14, the enrolled patients were classified into two groups of low P2RY14 expression and high P2RY14 expression, and the P2RY14 high-expression group had a substantially better OS than the P2RY14 low-expression group (P<0.05). Furthermore, we found that pathological stage and differentiation degree between the high and low expression groups of P2RY14 manifested significant statistical differences (P<0.05). P2RY14 expression was found to be an independent risk factor for the prognosis of lung cancer patients (P=0.000) in Cox regression analysis. The clinical data of the patients in our study were consistent with the data pertaining to LUAD in the database, indicating that our choice of P2RY14 as a predictive therapeutic target for LUAD was reasonable. Finally, Cox regression analysis was employed for determining the independent factors affecting the prognosis of lung cancer patients, and R language editing was used to create a nomogram on this basis. The nomogram’s C-index was 0.774 during internal validation (95% CI: 0.744–0.804), revealing the model has a high degree of accuracy in predicting the 5-year survival rate of LUAD patients, and has relevance as a reference and guide in clinical practice for screening and improving the prognosis of patients.
Conclusions
While the current work adds to our understanding of the link between P2RY14 and lung cancer, it does have certain limitations. First, when we looked at the link between P2RY14 and immune infiltration in LUAD patients, there was no explanation for immunological analysis based on subgroups such as preinvasive adenocarcinoma, microinvasive adenocarcinoma, invasive adenocarcinoma, and invasive adenocarcinoma variants. Second, while IHC revealed P2RY14 had an elevated expression in the cytoplasm of lung cancer cells, further analysis is required to probe into its molecular mechanism and role in tumor growth, metastasis, immune infiltration, and escape. Third, most of the analyses in the database were based on P2RY14 mRNA levels, and data will be more persuasive after further study based on protein levels. Finally, the diagnostic and prognostic significance of P2RY14’ in other pathological classes of lung cancer, such as large cell lung cancer and small cell lung cancer, was not investigated in the present work. Overall, the obtained data indicated that P2RY14 could be exploited as a possible novel predictive biomarker for lung carcinoma. Furthermore, based on the database and our own clinical research, we discovered that P2RY14 is substantially connected to immune infiltration, and it may have an essential involvement in limiting the immunological escape of tumor cells in the microenvironment of LUAD. As a result, our findings have the potential to advance understanding of not just P2RY14’s role in LUAD prognosis and immunotherapy, but also its translational use in LUAD prognosis and immunotherapy.
Acknowledgments
Funding: This study was supported by Nantong Municipal Health Commission (No. MSZ19234).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-115/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-115/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-115/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Nanjing Chest Hospital (No. 2021-KY094-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am 2016;25:447-68. [Crossref] [PubMed]
- Okamoto S, Togo S, Nagata I, et al. Lung adenocarcinoma expressing receptor for advanced glycation end-products with primary systemic AL amyloidosis: a case report and literature review. BMC Cancer 2017;17:22. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Wu GX, Raz DJ. Lung Cancer Screening. Cancer Treat Res 2016;170:1-23. [Crossref] [PubMed]
- Kron A, Scheffler M, Heydt C, et al. Genetic Heterogeneity of MET-Aberrant NSCLC and Its Impact on the Outcome of Immunotherapy. J Thorac Oncol 2021;16:572-82. [Crossref] [PubMed]
- Hayford CE, Tyson DR, Robbins CJ 3rd, et al. An in vitro model of tumor heterogeneity resolves genetic, epigenetic, and stochastic sources of cell state variability. PLoS Biol 2021;19:e3000797. [Crossref] [PubMed]
- Nicoś M, Krawczyk P, Crosetto N, et al. The Role of Intratumor Heterogeneity in the Response of Metastatic Non-Small Cell Lung Cancer to Immune Checkpoint Inhibitors. Front Oncol 2020;10:569202. [Crossref] [PubMed]
- Gregorc V, Lazzari C, Mandalá M, et al. Intratumoral Cellular Heterogeneity: Implications for Drug Resistance in Patients with Non-Small Cell Lung Cancer. Cancers (Basel) 2021;13:2023. [Crossref] [PubMed]
- Abbracchio MP, Burnstock G, Verkhratsky A, et al. Purinergic signalling in the nervous system: an overview. Trends Neurosci 2009;32:19-29. [Crossref] [PubMed]
- Chambers JK, Macdonald LE, Sarau HM, et al. A G protein-coupled receptor for UDP-glucose. J Biol Chem 2000;275:10767-71. [Crossref] [PubMed]
- Lazarowski ER, Harden TK. UDP-Sugars as Extracellular Signaling Molecules: Cellular and Physiologic Consequences of P2Y14 Receptor Activation. Mol Pharmacol 2015;88:151-60. [Crossref] [PubMed]
- Okada SF, Zhang L, Kreda SM, et al. Coupled nucleotide and mucin hypersecretion from goblet-cell metaplastic human airway epithelium. Am J Respir Cell Mol Biol 2011;45:253-60. [Crossref] [PubMed]
- Marucci G, Dal Ben D, Lambertucci C, et al. The G Protein-Coupled Receptor GPR17: Overview and Update. ChemMedChem 2016;11:2567-74. [Crossref] [PubMed]
- Shah K, Moharram SA, Kazi JU. Acute leukemia cells resistant to PI3K/mTOR inhibition display upregulation of P2RY14 expression. Clin Epigenetics 2018;10:83. [Crossref] [PubMed]
- Curet MA, Watters JJ. P2Y14 receptor activation decreases interleukin-6 production and glioma GL261 cell proliferation in microglial transwell cultures. J Neurooncol 2018;137:23-31. [Crossref] [PubMed]
- Woods LT, Forti KM, Shanbhag VC, et al. P2Y receptors for extracellular nucleotides: Contributions to cancer progression and therapeutic implications. Biochem Pharmacol 2021;187:114406. [Crossref] [PubMed]
- Musika W, Kamsa-Ard S, Jirapornkul C, et al. Lung Cancer Survival with Current Therapies and New Targeted Treatments: A Comprehensive Update from the Srinagarind Hospital-Based Cancer Registry from (2013 to 2017). Asian Pac J Cancer Prev 2021;22:2501-7. [Crossref] [PubMed]
- Cheng WC, Chang CY, Lo CC, et al. Identification of theranostic factors for patients developing metastasis after surgery for early-stage lung adenocarcinoma. Theranostics 2021;11:3661-75. [Crossref] [PubMed]
- Zhang S, Chen Z, Shi P, et al. Downregulation of death receptor 4 is tightly associated with positive response of EGFR mutant lung cancer to EGFR-targeted therapy and improved prognosis. Theranostics 2021;11:3964-80. [Crossref] [PubMed]
- Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013;3:1355-63. [Crossref] [PubMed]
- Rosenthal R, Cadieux EL, Salgado R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 2019;567:479-85. [Crossref] [PubMed]
- Song X, Chen Q, Wang J, et al. Clinical and prognostic implications of an immune-related risk model based on TP53 status in lung adenocarcinoma. J Cell Mol Med 2022;26:436-48. [Crossref] [PubMed]
- Shi A, Wang J, Wang Y, et al. Predictive value of multiple metabolic and heterogeneity parameters of 18F-FDG PET/CT for EGFR mutations in non-small cell lung cancer. Ann Nucl Med 2022; [Epub ahead of print]. [Crossref] [PubMed]
- Huo KG, Notsuda H, Fang Z, et al. Lung Cancer Driven by BRAFG469V Mutation Is Targetable by EGFR Kinase Inhibitors. J Thorac Oncol 2022;17:277-88. [Crossref] [PubMed]
- Jones GD, Caso R, Tan KS, et al. KRAS G12C Mutation Is Associated with Increased Risk of Recurrence in Surgically Resected Lung Adenocarcinoma. Clin Cancer Res 2021;27:2604-12. [Crossref] [PubMed]
- Kim HJ, Chae HZ, Kim YJ, et al. Preferential elevation of Prx I and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell Biol Toxicol 2003;19:285-98. [Crossref] [PubMed]
- Chua PJ, Lee EH, Yu Y, et al. Silencing the Peroxiredoxin III gene inhibits cell proliferation in breast cancer. Int J Oncol 2010;36:359-64. [PubMed]
- Mutharasu G, Murugesan A, Konda Mani S, et al. Transcriptomic analysis of glioblastoma multiforme providing new insights into GPR17 signaling communication. J Biomol Struct Dyn 2020; Epub ahead of print. [Crossref] [PubMed]
- Liu X, Shang X, Li J, et al. The Prognosis and Immune Checkpoint Blockade Efficacy Prediction of Tumor-Infiltrating Immune Cells in Lung Cancer. Front Cell Dev Biol 2021;9:707143. [Crossref] [PubMed]
- Ho KH, Chang CJ, Huang TW, et al. Gene landscape and correlation between B-cell infiltration and programmed death ligand 1 expression in lung adenocarcinoma patients from The Cancer Genome Atlas data set. PLoS One 2018;13:e0208459. [Crossref] [PubMed]
(English Language Editor: B. Draper)