Mycoplasma pneumoniae and Streptococcus pneumoniae caused different microbial structure and correlation network in lung microbiota
Background
Pneumonia is one of the most common illness in children, with high morbidity and mortality (1). Previous reports implicated that bacterial pathogens were the main contributors to pneumonia incidence and progression (2), and polymicrobial interaction was demonstrated, including Mycoplasma pneumoniae (M. pneumoniae), Chlamydophila pneumoniae (C. pneumoniae), Legionella pneumophila (L. pneumophila), Candida albicans (C. albicans), Pseudomonas aeruginosa (P. aeruginosa) and Staphylococcus aureus (S. aureus) (2).
Besides above-mentioned pathogens, lung was home to many kinds of bacteria (3,4). However, most of microbes in lung microbiota could not be cultured through conventional clinical detection. This will make it difficult to get the whole-picture of microbial interaction in the lung, and decrease the cure rate when uncultured or unknown lung pathogens existed. Culture-independent sequencing technology had been proved feasible in microbiota research on ventilator-associated, HIV-infected and idiopathic interstitial pneumonia (5-8). However, there was little report on microbial correlation network in microbiota at BALF, making it difficult to discern between bacterial carriage and infection.
Considering above-mentioned background, we conducted BALF sampling on 34 children with tracheomalacia or pneumonia and performed 16S rDNA analysis for 32 samples. Several issues that we intended to resolve were: (I) discrepancy of bacterial structure in children with tracheomalacia and pneumonia infected with different pathogens? (II) If C, D1 and D2 groups harbored discrepant bacterial correlation and how? This will improve understanding involvement of lung microbiota in pneumonia etiology, and distinct microbial correlation under pneumonia infected with S. pneumoniae and M. pneumoniae. Based on these findings, further analysis of mechanisms behind these correlations will promote more precise diagnosis and treatment of pneumonia.
Methods
Sampling, DNA preparation and sequencing
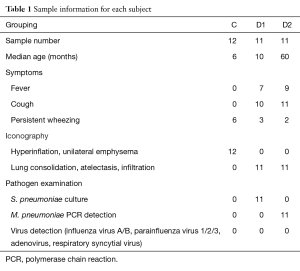
Parents of all children patients wrote the consent and this study was also approved by the Ethical Committee of Shenzhen Children’s Hospital under approval number 20150409436. Patients who fulfilled the following criteria were selected in this study: diagnosed with tracheomalacia, M. pneumoniae or S. pneumoniae pneumonia with characteristic chest radiographic abnormalities, patient symptoms, and clinical laboratory data (detailed in Table 1). BALF was collected by fiberoptic bronchoscopy and stored at −80 °C.
Full table
Microbial DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to manufacturer’s protocols. DNA detection, amplification of 16S rDNA V3-V4 region and sequencing was performed as prior report (9). Raw reads were deposited in NCBI Sequence Read Archive (SRA) database (Accession Number: SRP067201).
Data processing and bioinformatics analysis
Raw data was filtered using QIIME (version 1.17) (10): (I) the reads were removed if successive 50 bases held <20 quality score; (II) filtered reads with >2 mismatched bases in primer were also removed; (III) paired-end reads with >10 bases overlap were selected to assemble tags. Removing redundancy of assembled tags, species annotation, structural diversity and comparative analysis among three groups was conducted following previous report (11). Bacterial taxa with relative abundance >0.1% were selected to evaluate microbial correlation. The correlation graph was produced by Cytoscape (v3.3.0) under >0.9 Pearson index (12).
Results
Sample information and grouping
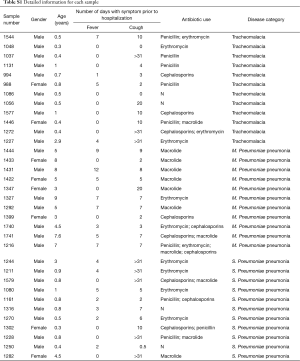
Totally 12 subjects had tracheomalacia (C group), 11 children were diagnosed to be infected by S. pneumoniae (D1 group) and other 11 with severe pneumonia were infected with M. pneumoniae (D2 group). Two samples were removed from subsequent analysis due to abnormal microbial structure, dramatically enriched by 44% Mycoplasma and 92% Haemophilus respectively. Other 32 samples were summarized in Table S1.
Full table
Microbial structure is discrepant among three groups
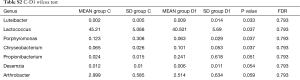
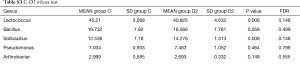
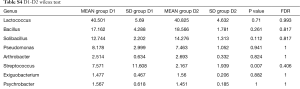
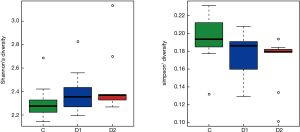
Totally 14,568 clean tags were produced for each sample on average, ranging from 9,383 to 18,946. At the phylum level, Firmicutes and Proteobacteria accounted for more than 90% in all samples, except the sample 1,037 which held 27.17% unclassified phylum. Lactococcus, Bacillus, Solibacillus, Pseudomonas, Streptococcus, Arthrobacter, Psychrobacter and Exiguobacterium were the top eight genus in three groups (Tables S2-S4), representing more than 90% relative abundance. It was difficult to discriminate the three groups based on principal components analysis (PCA), but several genus were differentially distributed apparently (Figure 1). By comparison to C group, children from D1 group harbored more abundant Streptococcus (7.51%±11.61%) and Propionibacterium (0.24%±0.62%), less abundant Lactococcus (40.50%±5.69%), Neisseria (0.05%±0.14%) and Moraxella (almost 0) (P value was summarized in Tables S2-S4). Propionibacterium (0.19%±0.43%), Mycoplasma (0.67%±1.25%), Massilia (0.81%±2.42%) and Staphylococcus (0.26%±0.71%) were enriched in D2 group, when compared to C group (P value was summarized in Tables S2-S4). C group harbored lower microbial diversity by comparison to D1 and D2 group (Figure 2).
Full table
Full table
Full table

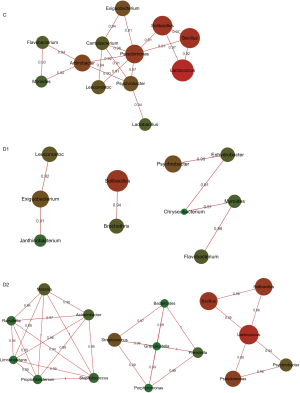
Different microbial correlation among C, D1 and D2 group
For children in C group, Pseudomonas and Arthrobacter were the core of correlation, with positive association to other normal colonizers, involving Bacillus, Solibacillus, Psychrobacter, Flavobacterium, Exiguobacterium and Lactobacillus, most of which were found to be the colonizers in the lung (4) [Figure 3 (C group)]. When children were infected with S. pneumoniae, the correlation network in C group was destructed, and Brochothrix, Janthinobacterium were involved in different correlation independently [Figure 3 (D1 group)]. The most complicated correlation was found in D2 group, and three strong correlation links were constructed [Figure 3 (D2 group)]. Enriched Staphylococcus, Propionibacterium and Massilia intended to build intensive association with Raoultella, Limnohabitans and Acinetobacter [Figure 3 (D2 group)]. Granulicatella mediated second network, involving Streptococcus, Porphyromonas, Bacteroides and Prevotella [Figure 3 (D2 group)]. The core of the third network was the most abundant Lactococcus, correlating positively with Bacillus, Pseudomonas, Solibacillus and Psychrobacter [Figure 3 (D2 group)].
Discussion
Increasing reports documented that respiratory tract is home to various microbes, which were associated with respiratory health (4). Sputum microbiota was demonstrated to be not concordant to microbes in the lung (13,14), and lung tissue could be collected only when lung transplantation will be conducted (3). This drove BALF sampling to be preferable strategy, in evaluating lung microbiota and corresponding involvement in lung diseases. We collected 32 samples with three symptoms: tracheomalacia, pneumonia infected with S. pneumoniae, pneumonia infected with M. pneumoniae. Previously identified microbial colonizers of lung, including Lactococcus, Pseudomonas, Prevotella, Fusobacteria, Veillonella, Haemophilus and Neisseria (15), were also found in this study, accounting for more than 90% of lung microbiota. Other pneumonia patients enriched bacterial species (5-8), including Staphylococcus and Streptococcus, were also increased in D1 and D2 group. Meanwhile, children with tracheomalacia harbored lower microbial diversity. This could be attributable to inhibitory effect of S. pneumoniae and M. pneumoniae on growth of naturally colonized microbes as well as more pathogen intrusion, and inter-individual difference which was demonstrated in intensive care unit (ICU) pneumonia patients (7).
The most interesting finding was that microbial correlation was totally different among three groups. In C group, Pseudomonas and Arthrobacter were in the core of interaction network and built positive relationship with other bacterial residents. Pseudomonas carriage was documented to be normal in prior reports (4), and this correlation also implicated homeostasis of microbiota at BALF, even if children from C group had tracheomalacia. When children were diagnosed pneumonia infected by S. pneumoniae, linkage of Pseudomonas and Arthrobacter to other bacterial colonizers were implicated to be broken up. This could be due to attachment of respiratory epithelial cells and evading host defenses by S. pneumoniae (16) through abundant various virulence factors like polysaccharide capsule and hydrogen peroxide, suggesting inhibitory effect of S. pneumoniae on other bacterial counterparts like S. aureus (17,18). Enrichment of S. pneumoniae in D1 group was also supported by clinical diagnosis based on traditional culture, and detection of M. pneumoniae in D2 group by single real time PCR, implicating accuracy of analysis results.
On the contrary, children infected by M. pneumoniae harbored more complicated bacterial correlation network. Besides Lactococcus and Granulicatella mediated network, D2 group enriched Staphylococcus, Propionibacterium and Massilia indicated to construct a complex positive interaction with Acinetobacter, Raoultella and Limnohabitans. However, Mycoplasma seemed to be outside of these relationships, possibly attributable to lack of antagonistic relationship between M. pneumoniae with other airway pathogens, and low immunity of pneumonia children in D2 group caused by M. pneumoniae indued severe symptoms (19). And S. aureus was proved to be the airway pathogen, interacting with S. pneumoniae, H. influenza and P. aeruginosa negatively (2), which could promote colonization of S. aureus at BALF of children infected with M. pneumoniae. However, Massilia will need more study to unravel its potential involvement in pneumonia inducement and progression, because there were just several reports about Massilia contribution to eye and skin illness (20).
Previous reports indicated that S. pneumoniae, S. aureus, P. aeruginosa and H. influenzae were also the main pathogenic agents for and accumulated in children with respiratory diseases (2-4). In this study, Pseudomonas and Haemophilus showed no discrepancy among three groups. We could not know if it was because children in C group were not totally healthy, or most of previous reports conducted different sampling. Other limitations of this study were insignificant discrepancy of some genera, including Porphyromonas and Psychrobacter, and ambiguous contribution of antibiotic, gender, age to difference among three groups. However, bacterial correlation in these groups will provide new insights into etiology of tracheomalacia and pneumonia infected with different bacterial pathogens.
To promote clinical application of these findings, several issues will be considered in our next step. Firstly, larger cohort study will be performed, and animal model will be collected to confirm and elucidate interaction network and mechanisms behind it. Then we will test if this could improve discerning between bacterial infection and carriage, with the aid of these correlation networks. Collectively, these findings will be applied in making more precise diagnosis and treatment on CAP, to lower the mortality and avoid overuse of antibiotic, including first-line macrolides (16).
Acknowledgements
Funding: This work was supported by the Key Medical Disciplines Building Project of Shenzhen (201506053).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Rudan I, O'Brien KL, Nair H, et al. Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health 2013;3:010401. [PubMed]
- Cillóniz C, Civljak R, Nicolini A, et al. Polymicrobial community-acquired pneumonia: An emerging entity. Respirology 2016;21:65-75. [Crossref] [PubMed]
- Adami AJ, Cervantes JL. The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2015;95:651-8. [Crossref] [PubMed]
- Cui L, Morris A, Huang L, et al. The microbiome and the lung. Ann Am Thorac Soc 2014;11 Suppl 4:S227-32. [Crossref] [PubMed]
- Toma I, Siegel MO, Keiser J, et al. Single-molecule long-read 16S sequencing to characterize the lung microbiome from mechanically ventilated patients with suspected pneumonia. J Clin Microbiol 2014;52:3913-21. [Crossref] [PubMed]
- Iwai S, Fei M, Huang D, et al. Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol 2012;50:2995-3002. [Crossref] [PubMed]
- Bousbia S, Papazian L, Saux P, et al. Repertoire of intensive care unit pneumonia microbiota. PLoS One 2012;7:e32486. [Crossref] [PubMed]
- Friaza V. Metagenomic analysis of bronchoalveolar lavage samples from patients with idiopathic interstitial pneumonia and its antagonic relation with Pneumocystis jirovecii colonization. J Microbiol Methods 2010;82:98-101. [Crossref] [PubMed]
- Schnorr SL, Candela M, Rampelli S, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 2014;5:3654. [Crossref] [PubMed]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. [Crossref] [PubMed]
- Moeller AH, Li Y, Mpoudi Ngole E, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A 2014;111:16431-5. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Cabrera-Rubio R, Garcia-Núñez M, Setó L, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol 2012;50:3562-8. [Crossref] [PubMed]
- Goddard AF, Staudinger BJ, Dowd SE, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A 2012;109:13769-74. [Crossref] [PubMed]
- Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011;184:957-63. [Crossref] [PubMed]
- Feldman C, Anderson R. Recent advances in our understanding of Streptococcus pneumoniae infection. F1000Prime Rep 2014;6:82. [Crossref] [PubMed]
- Bogaert D, van Belkum A, Sluijter M, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 2004;363:1871-2. [Crossref] [PubMed]
- Veenhoven R, Bogaert D, Uiterwaal C, et al. Effect of conjugate pneumococcal vaccine followed by polysaccharide pneumococcal vaccine on recurrent acute otitis media: a randomised study. Lancet 2003;361:2189-95. [Crossref] [PubMed]
- Fischer JE, Steiner F, Zucol F, et al. Use of simple heuristics to target macrolide prescription in children with community-acquired pneumonia. Arch Pediatr Adolesc Med 2002;156:1005-8. [Crossref] [PubMed]
- Chiquet C, Boisset S, Pechinot A, et al. Massilia timonae as cause of chronic endophthalmitis following cataract surgery. J Cataract Refract Surg 2015;41:1778-80. [Crossref] [PubMed]


