The expression of SALL4 is significantly associated with EGFR, but not KRAS or EML4-ALK mutations in lung cancer
Introduction
Lung cancer is the most common type of cancer and is the leading cause of cancer-related deaths worldwide. An ever increasing development of detection and treatment exists; also an increasing number of lung cancer patients are diagnosed in early stage and have access to surgical resection. However, the prognosis is still very poor, for the 5-year survival rate is less than 20% worldwide, this 5-year rate is about 18% in China and even less than 10% in some other Asian countries (1). An overview of the various types of lung cancer can help clarify the various dimensions of this critically serious disease. There are two major types of lung cancer: (I) non-small cell lung cancer (NSCLC) and (II) small cell lung cancer (SCLC). The NSCLC accounts for about 85% of lung cancer diagnoses; in addition, this type of cancer contains three histologic types: adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma (LCC) (2).
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are typically used as the first-line treatment in the NSCLC patients who are found to harbor the EGFR mutation. EGFR mutations mainly occur in exon 18 to 21, which encode the tyrosine kinase domain, more mutations occur in AC, especially in non-smoking Asian females (3). EML4-ALK and ROS1 are a two other genes of target therapy of lung cancer (4,5). Although target therapy could significantly increase survival and decrease undesirable side effects, drug resistant is often present. To improve the treatment of lung cancer, hopefully, more candidate genes which could be potential targets can be identified and used to both diagnose and offer more effective therapy.
Another factor in cancer, spalt-like transcription factor 4 (SALL4) (which is also known as ZNF797) belongs to the spalt family of zinc finger transcription factor and is required for early embryonic development (6,7). The most important function of SALL4 is to maintain the properties of embryonic stem cells (ESCs) by interacting with other important molecules, such as Oct4 and Nanog (8-10). SALL4 is required for DNA damage response in ESCs; it also maintains genomic stability during the expansion of ESCs (11). In human cancers, SALL4 is also overexpressed, such as acute myelocytic and lymphocytic leukemia’s, gastric cancer (12), glioma (13), and as well as in liver cancer. Up-regulation of SALL4 associated with poor prognosis in many cancers (12,13), and SALL4 expression were significantly correlated with gastric cancer cell metastasis to lymph nodes, especially in moderately differentiated tumor samples (14). Some investigators pointed out that serum SALL4 could be used as a new biomarker for early cancer detection (15), tumor recurrence, and poor survival (16). SALL4 is a potential, novel therapeutic target, but so far few investigations of SALL4 focus on lung cancer, even in the early clinical stages (17). Further review has shown that SALL4 expression is also significantly associated with drug-resistant. Up-regulated, the expression of SALL4 could decrease sensitivity to anti-cancer drugs, such as cisplatin, carboplatin, and paclitaxel; this SALL4 expression also may be involved in the recurrence of lung cancer after adjuvant chemotherapy (18). Further, no studies were found on the SALL4 expression and driver genes mutation. This study was conducted to investigate the relationship between the expression of SALL4 and driver genes mutation in lung cancer; therefore, 450 histopathologically diagnosed cases with lung cancer and 11 non-cancer patients were collected in this current study. The focus was to test, examine, and analyze the mutation status of EGFR, KRAS, EML4-ALK, and expression of SALL4.
Methods
Patients
In this study, all 450 lung cancer specimens were fresh tumor tissues obtained from the surgeries of lung cancer patients, and 11 normal lung tissues which were obtained from non-cancer patients; all people voluntarily joined this study with informed consent. For each case, the following medical records were gathered: clinical history, age, gender, cytologic diagnosis, and any subsequent histologic follow-up, if available. TNM (tumor, node, and metastases) staging was performed according the American Joint Committee for Cancer, 7th ed. (AJCC) staging system (19). The histologic subtypes of all patients were assessed and reassessed by at least two lung pathologists according to the 2011 International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) international multidisciplinary classification of lung ADC (20). The First Affiliated Hospital of Zhengzhou University ethics committee approved the protocol.
EGFR and KRAS mutation analysis
Extraction of DNA was performed first from fresh, surgically resected tumor samples. Next, DNA isolation was carried out using the AmoyDx Tissue DNA kit (Amoydx, Xiamen, China) according to the manufacturer’s instructions. Then, a highly sensitive, real-time PCR-based AmoyDx EGFR 29 Mutation Detection Kit was used to accurately identify 29 EGFR mutations in exons 18–21. Similarly, AmoyDx KRAS 7 Mutation Detection Kit was used to detect 7 KRAS mutations in codon 12 and 13 [these two AmoyDx Detection Kits of tumor mutation are approved, respectively, by China Food and Drug Administration (CFDA) for clinical use in China and Conformite Europeenne (CE) marked for in vitro diagnostic (IVD) products used in Europe].
EML4-ALK rearrangement analysis
Extraction of RNA was performed first from fresh, surgically resected tumor samples. Next, RNA isolation was done with the AmoyDx Tissue RNA kit (Amoydx, Xiamen, China) according to the manufacturer’s instructions. Then, EML4-ALK rearrangement was detected with AmoyDx EML4-ALK Gene Detection Kit (Amoydx, Xiamen, China) according to the manufacturer’s instructions. Briefly, first strand cDNA was synthesized from RNA using moloney murine leukemia virus (M-MLV) reverse transcriptase (Amoydx, Xiamen, China). The total amount of RNA should be within 0.1–5 μg, and A260/A280 value should be between 1.9 and 2.1. Target gene sequence was specifically amplified by proprietary primers. The amount of target cDNA was measured after each cycle in the data capture phase using a fluorescent probe. The fusion status of each sample and reference gene expression status were indicated by the FAM fluorescent signal.
SALL4 expression detection
First strand cDNA was synthesized from 1 μg total RNA using M-MLV reverse transcriptase (Amoydx, Xiamen, China) and downstream primers of the quantificational real-time polymerase chain reaction (qRT-PCR) primers as the anchor primer. The qRT-PCR primers were designed using Beacon Designer 7 (Premier Biosoft International, Palo Alto, Calif., USA). The primers of SALL4 are designed across introns. The reaction mixtures were incubated at 95 °C for 5 min followed by 42 °C for 1 h and 4 °C continuously.
The qRT-PCR reactions were performed using Mx3000P (Agilent, USA). The components of qPCR reaction buffer were 0.08 μL of SYTO9 working liquid, 25 mM of dNTPs, 10 mM of each primer, 2.5 mM of MgCl2, 5 U/µL of Taq DNA polymerase (Amoydx, Xiamen, China) in a total volume of 20 μL. The reaction mixture consisted of 2.5 µL of cDNA template. 18SrRNA was as internal reference.
The qRT-PCR protocol included an initial step of 95 °C for 5 min, followed by 45 cycles of 95 °C for 25 s and then annealed at 60 °C for 40 s, followed by one cycle of 95 °C for 1 min, 55 °C for 30 s, and 95 °C for 30 s. qPCR amplicons were subjected to melting curve analysis. The specificity of the qRT-PCR reactions was monitored with melting curve, analyzing by MxPro3000P and gel electrophoresis.
Statistical analysis process
The association of genes with clinical and pathologic characteristics was tested by Fisher’s exact test, and the independent t test was applied to continuous data. All statistical analysis was performed by using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA). All P values were based on a two-sided hypothesis. The statistical significance was set at P<0.05 for all analyses.
Results
Clinical characteristics and driver genes mutation status of samples
In order to investigate the association between driver genes mutation and expression of SALL4, we recruited 450 cases histopathologically diagnosed with lung cancer to carry out this study. We have listed the clinical characteristics of the patients (Table 1). More males were in the study, and the mean age is 61.5±9.2; more patients were over 61 years old. About 62.7% of the samples were in grade I, and about 20% of the patients were diagnosed as “positive” in their lymph nodes. The largest type we collected was ADC, followed by SCC. The two types accounted for about 82.4% of lung cancer samples. The status of EGFR, KRAS, and EML4-ALK were also listed (Table 1). The mutation rate of EGFR is about 47.7% in all lung cancer patients. L858R and 19-del are the main mutation type of EGFR, and account for about 23.6% and 18.0%, respectively. The mutation rate of KRAS and EML4-ALK, collectively, amounted to about 5%.
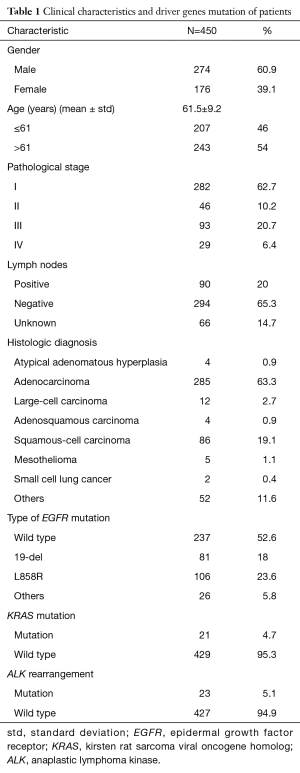
Full table
Clinical characteristics and driver genes mutation status in EGFR mutation samples
Because of increased mutations in EGFR, clinical information of EGFR mutation group was collected (Table 2). More females demonstrated EGFR mutation, accounting for 56.8%. The mean age of patients who harbored EGFR mutations was 60.9±9.3. About 67.6% patients were grade I, and about 21.6% patients were diagnosed as “positive” in their lymph nodes. About 87.3% EGFR mutations were in ADC and 2.3% in SCC. The results also showed that (I) two patients harbored both EGFR mutation and KRAS mutation; (II) eight patients carried both EGFR mutation and EML4-ALK rearrangements.
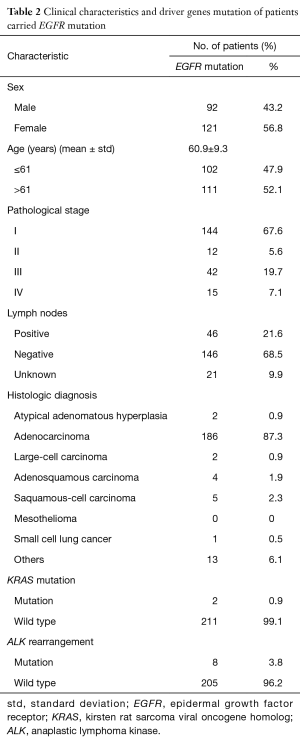
Full table
Association between SALL4 expression and driver gene mutation
In order to analyze the association between driver gene mutation and the expression of SALL4, specimens were collected from 450 lung cancer patients and 11 controlled group patients to determine the mutation status of EGFR, KRAS, EML4-ALK and the expression of SALL4 (Figure 1). Compared to the control group, the expression of SALL4 in patients who harbored driver genes mutation was higher, but only EGFR mutation presented significant difference (P<0.05).
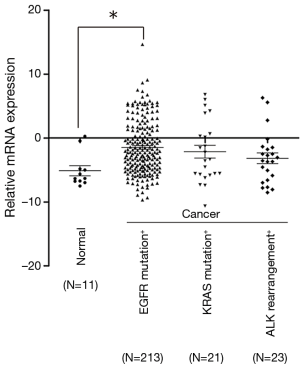
Association between SALL4 expression and EGFR mutation
In order to further investigate the association between the expression of SALL4 and EGFR mutation, two major mutation types were selected, L858R and 19-del, and a rare type which carried both L858R and 19-del mutation (Figure 2). Significant differences occurred between controls and cases in the expression of SALL4. The higher expression was present in all types of EGFR, including wild type, L858R, 19-del, and both mutations (P<0.05). An interesting result was that the expression of SALL4 in the patients who harbored two mutations was much higher than other mutation types of EGFR (P<0.05); however, no significant differences between other mutation types surfaced in the results.
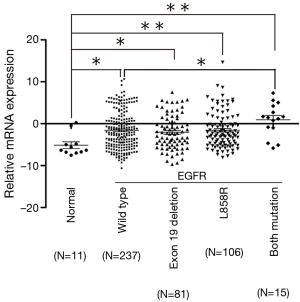
Discussion
Lung cancer is the most common type of cancer, and is the leading cause of cancer-related deaths worldwide. Finding a useful, therapeutic target is very meaningful for lung cancer treatment. SALL4 is considered a potential therapeutic target in cancer, but our review found few investigations on lung cancer. In the current study, we analyzed the association between SALL4 expression and mutant status of driver genes, including EGFR, KRAS, and EML4-ALK. The results showed that SALL4 is overexpressed in lung cancer, and the expression was significantly associated with EGFR mutation.
The mutations of EGFR mainly located in exon 18–21, which encode the tyrosine kinase domain in lung cancer. The significant association between EGFR mutations and the treatment of NSCLC was first described in 2004 (21,22). The two most common mutations are L858R in exon 21 and deletion in exon 19. L858R is higher than 19-del, 21% and 19%, respectively (23). Our study showed similar results: L858R mutation is about 23.6%, 19-del is about 18%, and the mutation which contained both L858R and 19-del is about 3.3%. These two mutations are sensitive to TKIs (24). The mutant rate of EGFR is about 47.7% in lung cancer, and about 74.7% in ADC. In the mutation analysis, 56% of the female carried EGFR mutation, and about 87.3% of the mutation was in ADC. This is accordance with a previous report that the mutation of EGFR is present in never-smoker Asian female with ADC (3). KRAS mutation differs between a westerner and an Asian, for the occurrences of KRAS are about 21% and 5–11%, respectively (25). Our data showed that about 5% of the patients harbored KRAS mutation, which was similar to this report. In addition, EML4-ALK is another important target in lung cancer therapy (5). The rearrangement of EML4-ALK is about 4–7% in NSCLC (25). Our study also detected about 5% lung cancer patients carried EML4-ALK fusion gene. Our analysis found 0.9% patients carried both EGFR and KRAS mutations, and 3.8% patients carried EGFR/EML4-ALK mutations.
SALL4 is over-expressed in many cancers, such as lymphocytic leukemia, gastric cancer and glioma; however, few studies investigated this relationship to ones who had lung cancer. Some former studies pointed out that only a small proportion of lung cancer positively expressed SALL4 (26,27), and there were differences between ADC and SCC (26). Another study showed that about 93% of samples presented, there was a more than two-fold change of SALL4 expression, and SALL4 was highly expressed even in early clinical stages (17). Our study also showed that the expression of SALL4 is higher in tumor tissues than normal tissues, and even about 80% of samples expressed greater than a two-fold change; there was, however, no significant difference between ADC and SCC (data not shown).
The results of the analysis of the association of SALL4 expression with EGFR, KRAS and EML4-ALK mutations were striking; these showed that compared to normal tissues, overexpression of SALL4 was detected in tumor tissues which harbored EGFR, KRAS and EML4-ALK mutations. The less positive tumor tissues in KRAS and EML4-ALK demonstrated insignificant differences. However, the EGFR mutation group showed significant results. Further analysis made with a different EGFR mutation showed us that SALL4 is up regulated in cancer tissues. However, there were no significant differences between wild type, L858R, and 19-del group. The most significant result was found in tissues which harbored both L858R and 19-del mutations. We believe our results indicate a more ample supply of tissues is needed to further confirm the data.
Finally, the SALL4 is up-regulated in lung cancer specimens with EGFR mutation indicates that the expression of SALL4 may be relevant with EGFR mutation. This conclusion could provide a new insight to lung cancer therapy. Therefore, further investigation of this mechanism is deemed to be much worthwhile.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The First Affiliated Hospital of Zhengzhou University ethics committee (approval number: research-2015-20) approved the protocol. Written informed consent was obtained from all patients.
References
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977-1010. [Crossref] [PubMed]
- Tang ER, Schreiner AM, Pua BB. Advances in lung adenocarcinoma classification: a summary of the new international multidisciplinary classification system (IASLC/ATS/ERS). J Thorac Dis 2014;6:S489-501. [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Antoniu SA. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363(18):1693-703. Expert Opin Ther Targets 2011;15:351-3. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Kohlhase J, Schuh R, Dowe G, et al. Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics 1996;38:291-8. [Crossref] [PubMed]
- Al-Baradie R, Yamada K, St Hilaire C, et al. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet 2002;71:1195-9. [Crossref] [PubMed]
- Wu Q, Chen X, Zhang J, et al. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem 2006;281:24090-4. [Crossref] [PubMed]
- Zhang J, Tam WL, Tong GQ, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 2006;8:1114-23. [Crossref] [PubMed]
- Jiang J, Chan YS, Loh YH, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 2008;10:353-60. [Crossref] [PubMed]
- Xiong J, Todorova D, Su NY, et al. Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J Cell Biol 2015;208:513-20. [Crossref] [PubMed]
- Liu J, Wang L, Yang A, et al. Up-regulation of SALL4 associated with poor prognosis in gastric cancer. Hepatogastroenterology 2014;61:1459-64.
- Zhang L, Yan Y, Jiang Y, et al. The expression of SALL4 in patients with gliomas: high level of SALL4 expression is correlated with poor outcome. J Neurooncol 2015;121:261-8. [Crossref] [PubMed]
- Momenzadeh D, Rahmanzadeh S, Rezaei-Tavirani M, et al. ZNF797 plays an oncogenic role in gastric cancer. Genet Mol Res 2014;13:8421-7. [Crossref] [PubMed]
- Ardalan Khales S, Abbaszadegan MR, Abdollahi A, et al. SALL4 as a new biomarker for early colorectal cancers. J Cancer Res Clin Oncol 2015;141:229-35. [Crossref] [PubMed]
- Zhang X, Belkina N, Jacob HK, et al. Identifying novel targets of oncogenic EGF receptor signaling in lung cancer through global phosphoproteomics. Proteomics 2015;15:340-55. [Crossref] [PubMed]
- Kobayashi D, Kuribayashi K, Tanaka M, et al. Overexpression of SALL4 in lung cancer and its importance in cell proliferation. Oncol Rep 2011;26:965-70. [PubMed]
- Yanagihara N, Kobayashi D, Kuribayashi K, et al. Significance of SALL4 as a drug-resistant factor in lung cancer. Int J Oncol 2015;46:1527-34. [PubMed]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. [Crossref] [PubMed]
- Lee HJ, Lee CH, Jeong YJ, et al. IASLC/ATS/ERS International Multidisciplinary Classification of Lung Adenocarcinoma: novel concepts and radiologic implications. J Thorac Imaging 2012;27:340-53. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Wang X, Wang G, Hao Y, et al. A comparison of ARMS and mutation specific IHC for common activating EGFR mutations analysis in small biopsy and cytology specimens of advanced non small cell lung cancer. Int J Clin Exp Pathol 2014;7:4310-6. [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013;24:2371-6. [Crossref] [PubMed]
- Rodriguez E, Chen L, Ao MH, et al. Expression of transcript factors SALL4 and OCT4 in a subset of non-small cell lung carcinomas (NSCLC). Transl Respir Med 2014;2:10. [Crossref] [PubMed]
- Fujimoto M, Sumiyoshi S, Yoshizawa A, et al. SALL4 immunohistochemistry in non-small-cell lung carcinomas. Histopathology 2014;64:309-11. [Crossref] [PubMed]


